Commentary
Sea Star Asterias Rubens Fab Gene and Bioinformatic Data
2893
Views & Citations1893
Likes & Shares
The Asterias rubens Fab gene was isolated in 2016. Its transcriptome was analyzed in bioinformatics according the method of Marchler-Bauer. Identities mainly with other sea star genomes were repertoried in the precedent report.
INTRODUCTION
The objective of this work is to analyze Fab DNA sequence from the sea star Asterias rubens (Asterids, Echinodermata) which belong to Invertebrates [1].
Our Starting material is the following DNA sequence which is translated in 5’-3’
CAGAGTCACCAGGTGCTGTGTTGACATCAGCACTTTCTGAATCCACCTCCATTGGTTGATCCACTTCCTCTTTCACTATAGTTGACGATTTGATATCAACACTGTCACCTGTAAGCGCATCAACACCGCTTGTTATATCAACAGACGATCTCCTTTGTGGATCCACTTTTTGATCGGTGGATAGAGGCGGAGTGGATGACTCCATCTTTGCTAATGTTTTGTCATCGGCTAATGTTGATGAACTTTCTTCAGCTTTTACCTCGGTCTTCACATCCACTGTTTGGTTTTCTGATTTCACGGAAACAGTTGATGTCGTTTTGACTGGATCCACTGACTTATCAGCCAATGTTCCCTGCGGAGTTGTTGATGTACTTTGTTGAGAGATGGCTGTGAAGGGATTTGTGGCGGGATCGAAGGAAGCTCCTAAGCCTTCCTCCCCTGGGGTTGGCGTTTCTAGAAGCGAGGAATCTTTGGCATCTTCGAAACACAAGGGATGGTATGTCCTGGAGTTGAACCTGACAGCATCTCTCAGTTTCCAAACCTCTTCATCTTCATCAAAGAATTGCTCAAACGGCTCATGACACAGAGAACACATCTCATCGCCAGTGACGTTTCCATCAATAGCCACACAGAGGATGCCATCTTTATCAGGTTTAGACTCTGCAGTCATCTCCTCAAAGAAACTGCTCCTCTCTTCATCCACTTCGTTGATTTCTTCAAAGTCCAACCACTGATCGACTTCATAGAACCACTTCCTGGACTGAACAGCCTTCACTCCATCCTTCTCACGACGATTCTGACGGAAGTGCCAGTCCAGATGTTTCCGATATCTGTCCATTTCATCAATGACAAAGCGCATACCA
RESULTS
- Blastn original sequence Data base:
Standard data bases (nr.) have been used [2,3] and optimized for highly similar sequences (megablast)
The DNA molecule included a query length of 863
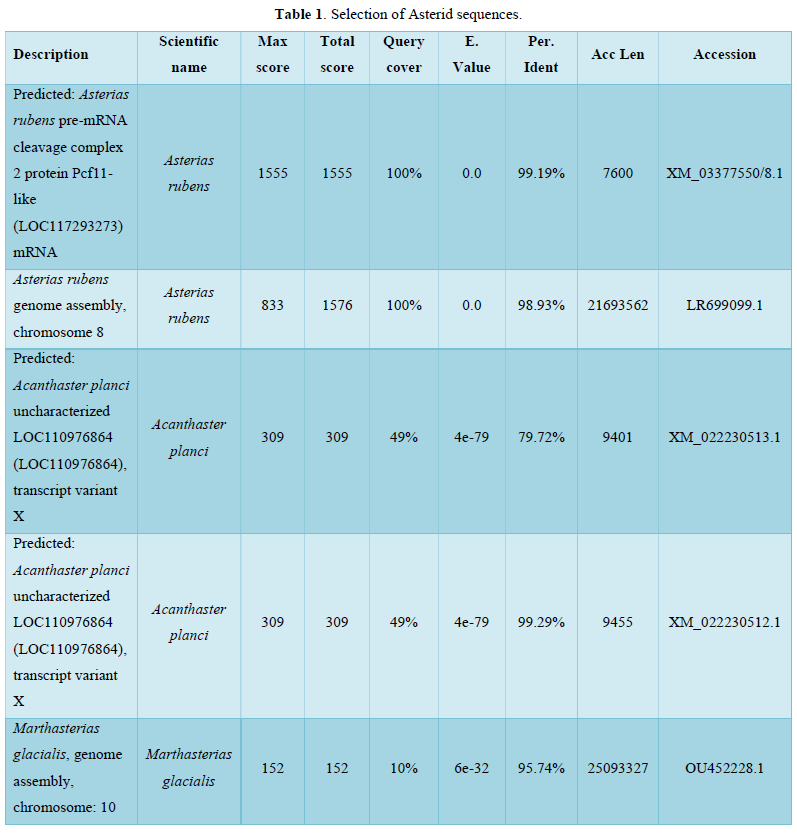
Among sequences producing significant alignments, 5 sequences were selected as shown in Table 1.
The graphic summary follows below Figure 1.
- BlastX original sequence:
As precedingly the Molecule type is DNA. The characteristics were the following:
- Query length: 863
- Genetic code: Standard
- Database: Non-redundant protein sequences (nr)
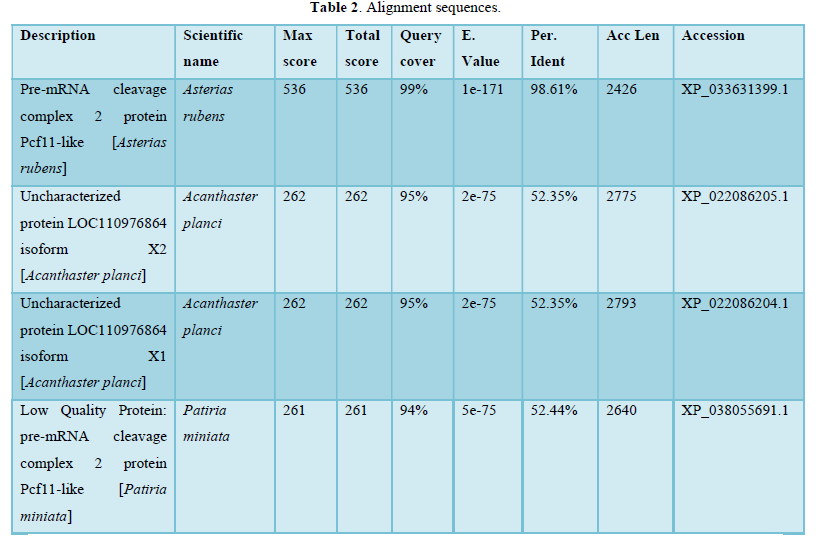
- Sequences producing significant alignments: more than 100 sequences were found as shown below in Table 2.
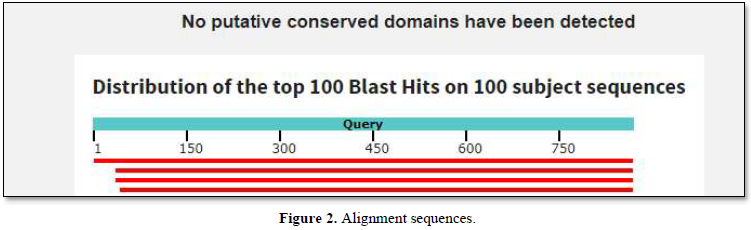
The graphic summary was resumed as shown below Figure 2.
Conclusion
No putative conserved domains as homologies with Ig domains have been found. Such homologies were detected with the Fc receptor gene from Asterias rubens [4].
In the present study alignment sequences present mainly analogies between Fc receptor genes and sophisticated proteins which belong especially to Asterias rubens (Protein Pcf11-like: 99, 19 %identities) or other starfishes at a less degree. Table 2 corroborates these same results.
We retain Fab gene and Fc receptor gene exist in Echinodermata as it was already demonstrated in 2016.





- Leclerc M, Kresdorn N (2016) Evidence of Fab Fragment Gene in an Invertebrate: The Sea Star Asterias. Int J Biotech and Bioeng 2(1): 37-38.
- Marchler-Bauer A, Bo Y, Han L, He J, Lanczycki CJ, et al. (2017) CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45(D1): D200-D203.
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, Lu S, Chitsaz F, et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res 43(D): D222-D226.
- Leclerc M (2022) IJBA submitted.



