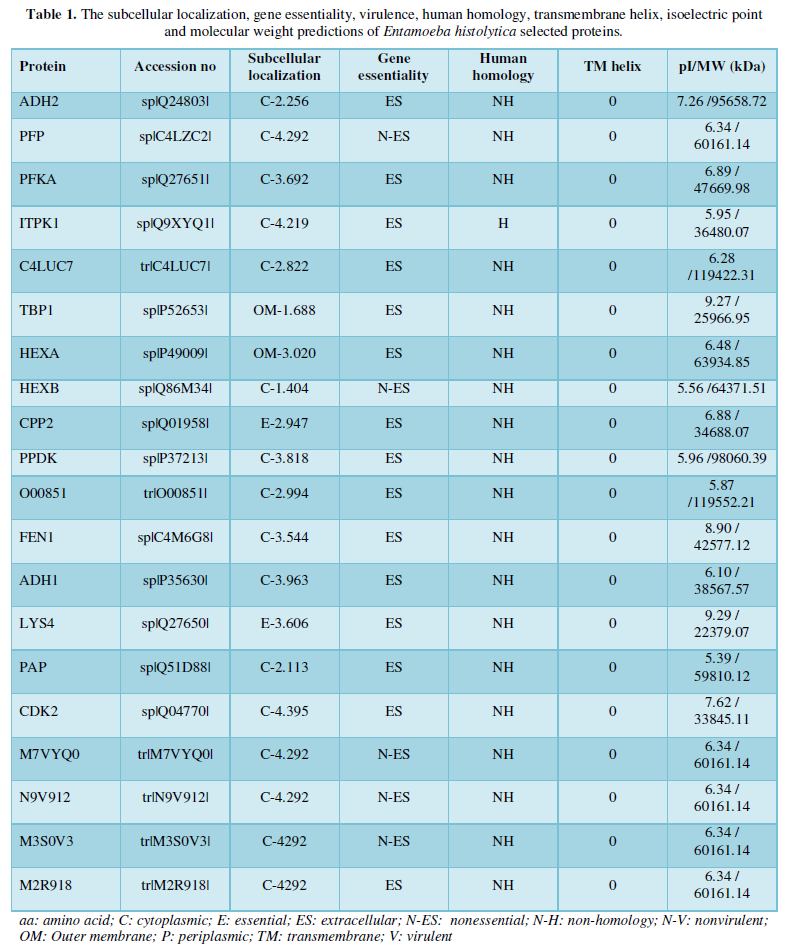
Research Article
A Bioinformatics Approach to Designing a Multi-Variant Vaccine Against Entamoeba histolytica
6658
Views & Citations5658
Likes & Shares
Entamoeba histolytica is the parasite that is responsible for amoebiasis and remains one of the very important best three parasitic causes of mortality worldwide. With increased travel and emigration to developed countries; such infection is becoming more common in non-endemic areas. Although the bulk of people infected with E. histolytica remains asymptomatic, some presents with amoebic colitis and disseminated disease. As more is learned about its pathogenesis and thus the host’s immune reaction, the potential for developing a vaccine holds promise. This narrative reported outline the present knowledge regarding E. histolytica insight within the development of a vaccine. During this study, immune informatics approach was used to design a completely unique epitope based oral vaccine against E. histolytica. This is due to the rise in resistance to antibiotics for the control, leading to an efficient vaccine as an alternative, which remains a challenge. Therefore, a rational, strategic, and efficient vaccine design against E. histolytica is important where the utilization of the foremost current bioinformatics tools like GEPTOP CELLO NCBI which some predictions results of which ADH2, PFP, PFKA, ITPK1, C4LUC7, HEXB, PPDK, O00851, FEN1, ADH1, PAP, CDK2, M7VYQ0, N9V912, M3S0V3 and M2R918 possessed a cytoplasmic location; LYS4and CPP2 were extracellular, whereas TBP1, and HEXA were predicted as outer-membrane proteins, none were inner-membrane and periplasmic. In this study, immune informatics approach was use to design a completely unique epitope based oral vaccine against E. histolytica. The vaccine designed fulfilled non-allergenicity, antigenicity, and appropriate solubility, relative molecular mass and isoelectric point.
Keywords: Immunoinformatics, Allergenicity, Entamoeba histolytica, Antigenicity, Multi-Variant -Vaccine
INTRODUCTION
Amoebiasis, or amoebic dysentery, could also be a term used to describe an infection caused by the protozoan Entamoeba histolytica [1]. Mostly infections were commonly known as asymptomatic, but invasive intestinal disease might occur manifested with several weeks of cramping, abdominal pain, watery or bloody diarrhea, and weight loss [1]. Disseminated, extra intestinal disease like liver abscess, pneumonia, purulent pericarditis, and also cerebral amoebiasis has been described [1]. Worldwide, it has been estimated that up to 50 million people are suffering from E. histolytica, primarily in developing countries, and it's liable for over 100,000 deaths a year [2]. Transmission generally occurs by the ingestion of infected water or food thanks to fecal excretion of cysts, and even fecal-oral transmission within household and through male homosexual activity [1,3,4]. Vaccines were currently being investigated in rodent and a non-human primate model appears promising. Using both native and recombinant kinds of the amoebic Gal/GalNAc lectin has been presented for protection against intestinal and liver infections [5]. Investigated a vaccine in baboon models, a natural host for E. histolytica [6]. Cholera toxin B subunit was co administered with Gal-lectin which resulted in significant, moderate level of protection against E. histolytica reinjection. The vaccinated baboons displayed higher titers of intestinal anti-peptide IgA, intestinal anti-lectin IgA, and also serum anti-peptide IgG antibodies, followed up colonoscopy showed no sign of inflammatory colitis or amoebic invasion [6]. Targeting other components of E. histolytica, like serine-rich protein and thus the 29-kDa- reductase antigen, has been shown successful against ALA in rodent models [5]. More rigorous testing for the event of a successful vaccine against E. histolytica is warranted. Apart from identifying target immunogenic proteins, the mixture of doses, adjuvants, and boosts must be optimized [7]. The responses in animal models, non-human primates especially [6], suggest that it's going to be possible to develop a viable human vaccine, advancing to urge beyond the preclinical stage. Sadly, however, the induction of LTM has not yet been presented in animal models, which is the hallmark of a successful vaccine.
The technological advances within the fields of genomics, proteomics, human immunology, and structural biology have provided the molecular information for the invention and prediction by bioinformatics tools of novel antigens, epitopes, and style of vaccines against pathogenic bacteria, like meningococcus B [8-10]. Bioinformatics tools can predict sequences with binding affinity to MHC alleles and epitopes of T and B cells [11]. The main target on vaccine design and development has changed to the assembly of peptides composed of multiple epitopes (multi epitope vaccines), supported linear arrangements, as a totally unique alternative. Additionally, epitope-based vaccines have demonstrated various advantages, including safety, the chance to rationally engineer the epitopes for increased potency, breadth, and antigenicity, and therefore the possibility to focus large repertories of immune responses on conserved epitope sequences [12]. This study was an in-silico attempt aimed toward designing a completely unique oral vaccine against E. histolytica was in silico designed.
METHODS
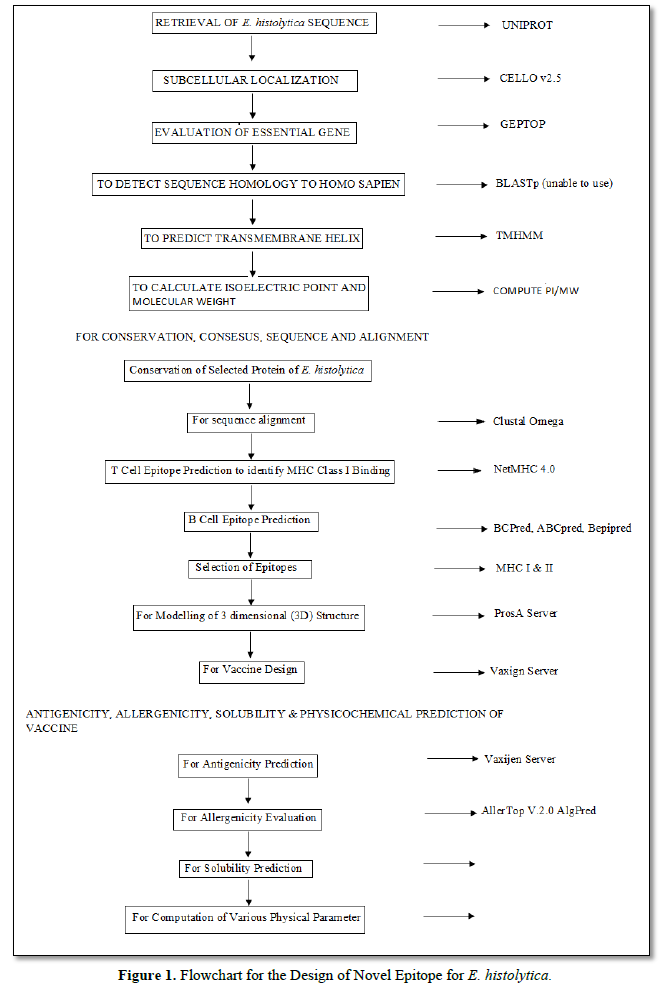
Selection and Antigenic Evaluation of Protein
- histolytica proteins were selected based on the following criteria: (1) reported antigenicity, (2) virulence, and (3) proteins related to the mechanisms of adhesion. The entire twenty protein sequences were retrieved from UniProt reference sequence database in FASTA format. For bacterial protein subcellular localization prediction, CELLO v2.5 was used [13]. The database of Gep Top was used to evaluate the essentiality of genes [14]. PATRIC3.5.16 database was used for study of the virulence role of proteins. Using BLASTp to screen proteins for detecting sequence homologs to Homo sapiens. Prediction of transmembrane (TM) helixes, TMHMM v2.0 was used. Compute pI/Mw tool was used to calculate the estimated isoelectric point and molecular weight of all amino acid sequences [15].
Phylogenetic Evolution
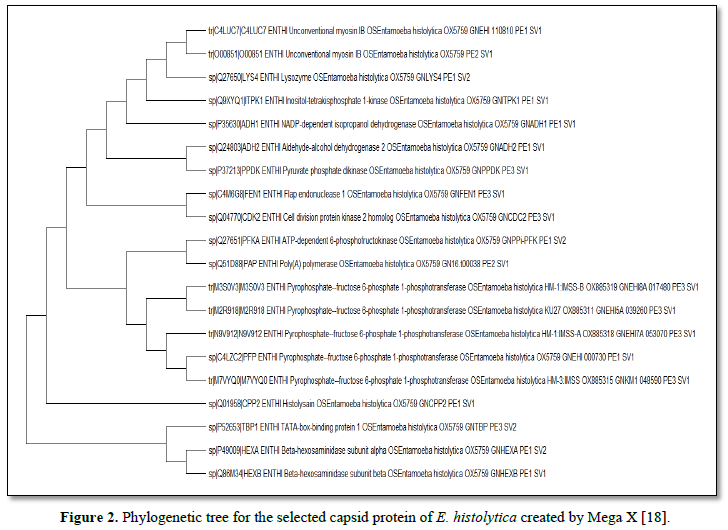
Phylogenetic tree of the retrieved sequence of the capsid protein of E. histolytica was created using Mega X software. 20 of the protein trees were constructed using maximum likelihood parameter in the software [16].
Conservation, Consensus Sequence and Alignment
Conservation of selected proteins was analyzed using E. histolytica strain 26695 as the reference. For proteins with high variability, a consensus sequence was generated using sequences of world E. histolytica representative strains (20 complete genome sequences). CLC Main Workbench v7.8 was performed for sequence alignment (QIAGEN Bioinformatics), Emboss Cons, and T-Coffee software.
T Cell Epitope Prediction
To identify MHC-I binding epitopes, NetMHC 4.0 server was used [17]. Fifty-one human leukocyte antigen (HLA) alleles (HLA-A, -B, -C, and -E) and six murine alleles (H-2) were evaluated. Base on the MCH-I binding epitopes predictions were calculated for nine-mers epitopes with a threshold for strong binders of 0.5% and a threshold for weak binders of 2%.
For MHC-II binding epitopes, NetMHCII 2.3 server; predictions which were obtained for 20HLA- DR alleles, 20 HLA-DQ, 9 HLA-DP, and 7 mouse H2 class II alleles using a threshold of -99.9, threshold for the strong binder of 5%, and threshold for the weak binder of 20%.
B Cell Epitope Prediction
Linear B cell epitopes of 20-mers were predicted utilizing ABCpred with a threshold of 0.7. The second was BCPred server which was applied with a specificity threshold of 75%. For BepiPred server, only amino acids with score >1.0 were considered for the downstream analysis [18].
Selection of Epitopes
They were selected based on the following criteria: (1) 20-mer epitopes, (2) epitopes matching on all algorithms, if possible, and (3) potential to bind with the maximum number of MHC-I and MHC-II alleles. Using Clustal Omega server for the selection of sequences that were aligned and overlapped.
Vaccine Design
Vaxign Server was used to get the vaccine design which shows the protein accession, gene symbol, localization probability, adhesion probability, transmembrane helices and the protein length [19].
Protein Prediction and Validation of Secondary and Tertiary Structures
The secondary structure of the multi-epitope antigen was predicted using PSIPREDv3.3 Protein Prediction and validation of secondary and tertiary structures [20]. The three-dimensional (3D) structure modeling was performed using Swiss-Model server. Jmol was used for visualizing 3D structures of proteins. For refinement of 3D model structure, Galaxy Refine and Galaxy Loop were applied [21].
The best model was validated by the ProSA web and ERRAT. The residue-by-residue stereochemical qualities of models were validated by Ramachandran plot obtained from PROCHECK server [22]. The best-refined model was selected.
Antigenicity, Allergenicity, Solubility and Physicochemical Predictions of Vaccine
For antigenicity prediction, VaxiJen server was used for antigenicity, allergenicity, solubility, and physicochemical predictions of vaccine. AllerTOP v.2.0 and AlgPred servers were used for allergenicity evaluation. For solubility prediction, SOLpro server was used. Finally, ProtParam allowed the computation of various physical and chemical parameters [15].
RESULTS AND DISCUSSION
Protein Selection and Evaluation
It has been shown that the combination of a long list of antigens do increase the effectiveness of vaccines [23] evaluated a multi component vaccine in human volunteers; they observed that the vaccine was safe but highly immunogenic, inducing long-lasting humoral and cellular responses to the antigens. Twenty 20 proteins were selected for this vaccine candidate. These were further screened for the next step for further parameters, including subcellular localization, essentiality, virulence, non-human homology, TM helixes, and molecular weight. Our predictions showed that sixteen: ADH2, PFP, PFKA, ITPK1, C4LUC7, HEXB, PPDK, O00851, FEN1, ADH1, PAP, CDK2, M7VYQ0, N9V912, M3S0V3 and M2R918 possessed a cytoplasmic location; LYS4 and CPP2 were extracellular, whereas TBP1, and HEXA were predicted as outer-membrane proteins as shown in Table 1. Both surface and extracellular proteins are good targets to develop a vaccine aiming toward prevention of bacterial infections and diseases [24].
In the analysis of essential genes by using GepTop package, fifteen (15) out of the 20 selected proteins were predicted to have essential genes. ADH2, PFKA, ITPK1, C4LUC7, TBP1, HEXA CPP2, PPDK, O00851, FEN1, ADH1, LYS4, PAP, CDK2 and M2R918 were identified to have essential genes. Essential genes are those genes for an organism that are critical for its survival; essential genes are of particular importance because of their theoretical and practical applications such as studying the robustness of a biological system, defining a minimal genome/organism and identifying effective therapeutic targets in pathogens [14]. Homology analysis of the 20 prioritized proteins using BLASTp revealed <60% identity, which was significant and important to declare the sequences as nonhuman homologs; A good vaccine targets should not be human homologues so as to avoid autoimmunity. The prediction of the topology of proteins by TMHMM showed that none of them have helix. Finally, molecular weights calculated by Compute pI/Mw tool of 4 proteins resulted to weigh <60,161.14 kDa respectively, also 2 protein resulted to weight (Table1).
Good vaccine candidates are considered as those that do not have similarity with human proteins in order to avoid or prevent the generation of a potential autoimmune response; they must also lack TM regions, to facilitate their expression. Other features of a good vaccine potential are that it should possess good antigenic properties, which are vital for disease pathogenesis and for protection against the antigen [25]. Some authors have used these approaches in the selection of candidate proteins for thein silico design of E. coli vaccines [26].
T and B Cell Epitopes
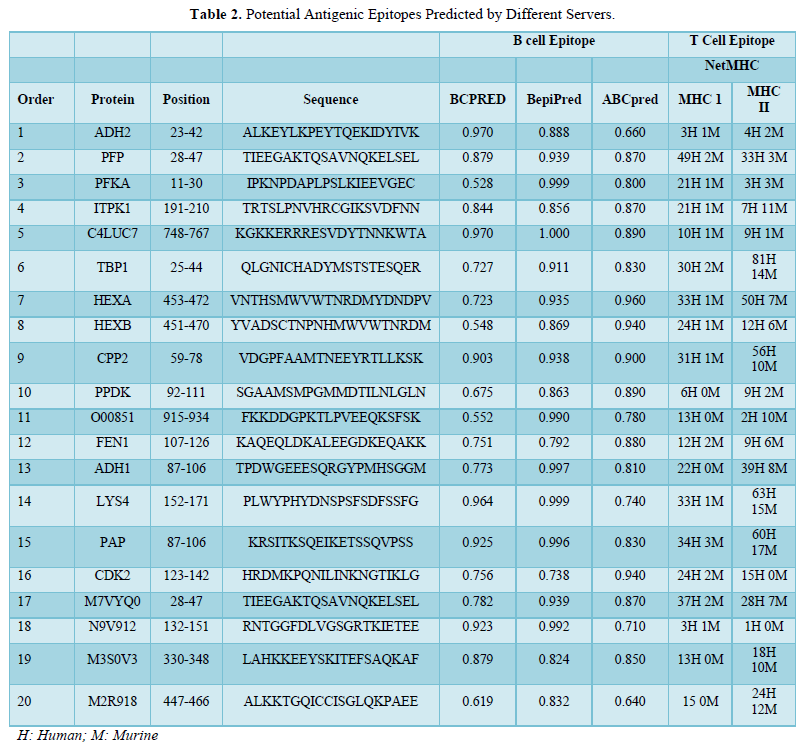
The prediction analysis by different bioinformatics servers for T and B cells (using MHC-I/-II alleles for human and mouse BALB/c) allowed the selection of 20 epitopes based on their score, number of alleles, and agreement between the servers used. Epitopes segment obtained were: ADH223-42, PFP28-47, PFKA11-30, ITPK1191-210, C4LUC7748-767, TBP125-44, HEXA453-472, HEXB451-470, CPP259-78, PPDK92-111, O00851915-934, FEN1107-126, ADH187-106, LYS4152-171, PAP87-106, CDK2123-142, M7VYQ028-47, N9V912132-151, M3S0V3330-348 and M2R918447-466 (Table 2 and Figure 1).
Protein Structure Prediction
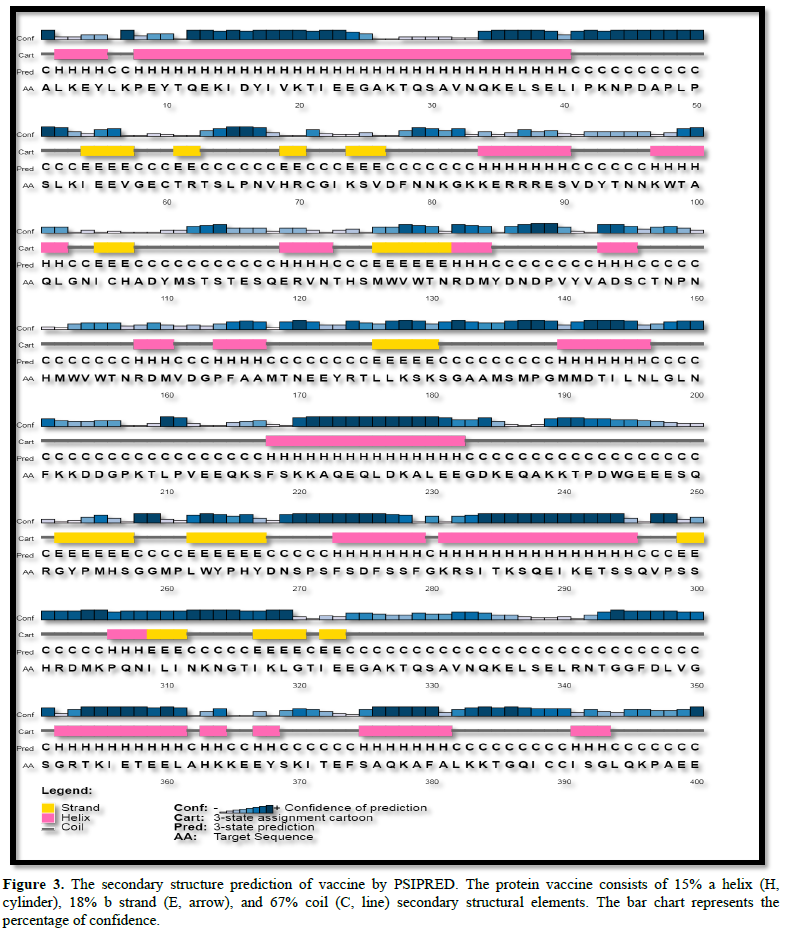
The vaccine is composed of 400 amino acids, and prediction of secondary structure showed that it contains 67% a helixes, 15% ꞵ sheets, and 18% others (random coil and ꞵ-turn), as shown in Figure 3.
Five 3D models of protein vaccine were generated among which the model with the highest c-score = 5 was selected for further refinement; the c-score range is typically from 5-6, the higher the value, the higher the confidence.
The quality and potential errors in the best model were analyzed. The initial input model z-score was-9.94, which falls within those commonly observed in similar size-native proteins (Figure 4A). ProSA-web indicated that the preliminary model requires refinement processes. Hence, the raw model was subjected to loop refinement and energy minimization using galaxy refine. After all refinement procedures, ERRAT factor was improved from 96.8 to 97.9. The z-score of the final model reached a value of -10.11


(Figure 4B). The starting model was given (Figure 5).
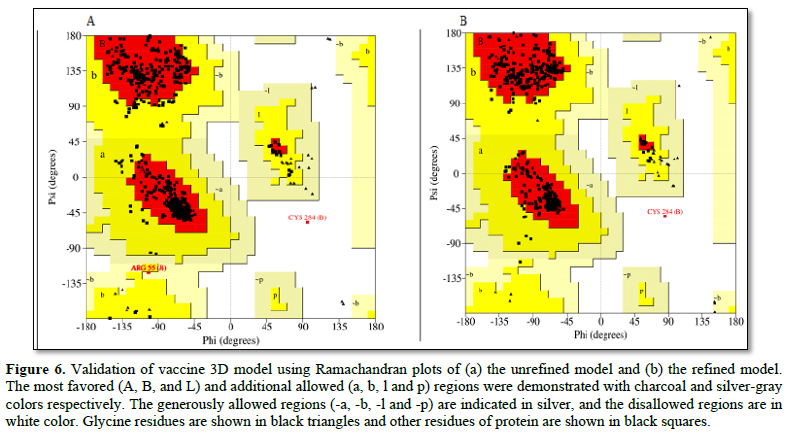
To validate the 3D models, Ramachandran plot analysis was performed before and after refinement processes. The Ramachandran plots of the unrefined model indicated that 88.1%of residues were located in most-favored regions, 11.3% in the additional allowed region, 0.4% in generously allowed regions, and 0.2% in disallowed regions of the plot (Figure 6A).
The refined model showed that 94.5% of residues were located in most-favored regions, 5.3% in additional allowed regions, 0.0%ingenerously allowed regions, and only 0.2%in disallowed regions (Figure 6B).
Antigenicity, Allergenicity, Solubility and Physicochemical Parameters of the Vaccine
An antigenicity score of 0.33722625 was obtained. The allergenicity prediction showed that the vaccine is allergenic. The molecular weight and theoretical pI of protein were 45189.22 kDa and 6.02, respectively. The final model was given (Figure 7).


CONCLUSION
In this study, we designed a novel multi epitopes oral vaccine against E. histolytica based on bioinformatics analysis to produce a huge and robust cellular and humoral immune response for E. histolytica prevention. This novel oral vaccine design could be a good vaccine candidate against E. histolytica. However, to make the therapeutic and prophylactic effect of our oral vaccine design valid, in vitro and in vivo immunological studies are required.
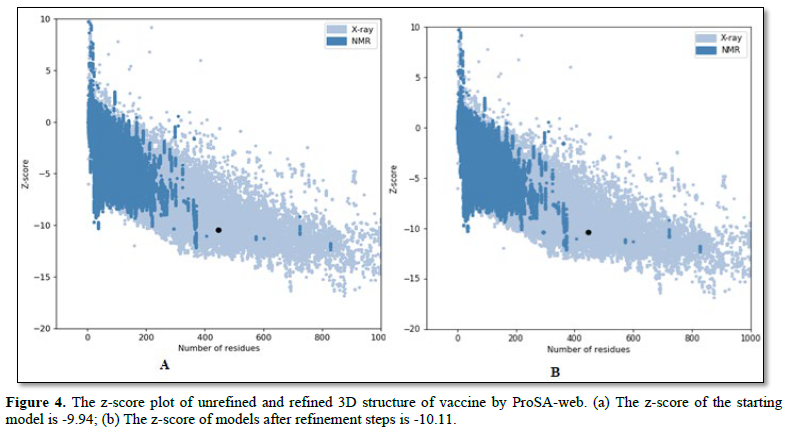


The z-scores indicates overall model quality and is depicted as a black spot. The z-scores of all experimentally determined protein chains in current protein data bank (PDB) from NMR spectroscopy (Charcoal) and X-ray crystallography (silver). 3D, three dimensional.



- Haque R, Huston CD, Hughes M, Houpt E, Petri WA (2003) Amebiasis. N Engl J Med 348(16): 1565-1573.
- Bercu TE, Petri W, Behm JW (2007) Amebic colitis: New insights into pathogenesis and treatment. Curr Gastroenterol Rep 9(5): 429-433.
- Cheepsattayakorn A, Cheepsattayakorn R (2014) Parasitic pneumonia and lung involvement. Biomed Res Int 2014: 874021.
- Salit IE, Khairnar K, Gough K, Pillai D (2009) A possible cluster of sexually transmitted Entamoeba histolytica: Genetic analysis of a highly virulent strain. Clin Infect Dis 49(3): 346-353.
- Quach J, St-Pierre J, Chadee K (2014) The future for vaccine development against Entamoeba histolytica. Hum Vaccin Immunother 10(6): 1514-1521.
- Abd Alla MD, Wolf R, White GL, Kosanke SD, Cary D, et al. (2012) Efficacy of a Gal-lectin subunit vaccine against experimental Entamoeba histolyticainfection and colitis in baboons (Papio sp.) Vaccine 30(20): 3068-3075.
- Simpson I, Woolley IJ, Gardiner BJ (2015) Caught in the act… a case of ruminant Amoebic colitis JMM Case Rep 2(4): 1-3.
- Serruto D, Bottomley MJ, Ram S, Giuliani MM, Rappuoli R (2012) The new multicomponent vaccine against meningococcal serogroup B, 4CMenB: Immunological, functional and structural characterization of the antigens. Vaccine 30: 87-97.
- O’Ryan M, Stoddard J, Toneatto D, Wassil J, Dull PM (2013) A multi-component meningococcal serogroup B vaccine (4CMenB): The clinical development program. Drugs 74: 15-30.
- Rappuoli R, Bottomley MJ, D'Oro U, Finco O, De Gregorio E (2016) Reverse vaccinology 2.0: Human immunology instructs vaccine antigen design. J Exp Med 213: 469-481.
- Rosa DS, Ribeiro SP, Fonseca SG, Almeida RR, Santana VC, et al. (2015) Multiple approaches for increasing the immunogenicity of an epitope-based anti-HIV vaccine. AIDS Res Hum Retroviruses 31(11): 1077-1088.
- Livingston B, Crimi C, Newman M, Higashimoto Y, Appella E, et al. (2002) A rational strategy to design multi-epitopes immunogens based on multiple Th lymphocyte epitopes. J Immunol 168(11): 5499-5506.
- Chin-Sheng Y, Yu-Ching C, Chih-Hao L, Jenn-Kang H (2006) Prediction of protein subcellular localization. Proteins 64: 643-651.
- Wei-Hua C, Lu G, Chen X, Xing-Ming Z, Bork P (2017) OGEE v2: An update of the online gene essentiality database with special focus on differentially essential genes in human cancer cell lines. Nucleic Acids Res 45(D1): D940-D944.
- Wilkins MR, Gasteiger E, Bairoch A, Sanchez JC, Williams KL, et al. (1999) Protein identification and analysis tools in the ExPASy server. Methods Mol Biol 112: 531-552.
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol Evol 35(6): 1547-1549.
- Andreatta M, Nielsen M (2016) Gapped sequence alignment using artificial neural networks: Application to the MHC class I system. Bioinformatics 32: 511-517.
- Jespersen MC, Peters B, Nielsen M, Marcatili P (2017) BepiPred-2.0: Improving sequence-based B-cell epitope prediction using conformational epitopes. Nucleic Acids Res 45: 24-29.
- Xiang Z, He Y (2013) Genome-wide prediction of vaccine targets for human herpes simplex viruses using Vaxign reverse vaccinology. BMC Bioinformatics 14(Supple 4): S2.
- McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16: 404-405.
- Park H, Ko J, Joo K, Lee J, Seok C, et al. (2011) Refinement of protein termini in template-based modeling using conformational space annealing. Proteins 79: 2725-2734.
- Laskowski RA, MacArthur MW, Thornton JM (2012) PROCHECK: Validation of protein-structure coordinates. Int Tables Crystallography F: 684-687.
- Rossi G, Ruggiero P, Peppoloni S, Pancotto L, Fortuna D, et al. (2004) Therapeutic vaccination against Helicobacter pylori in the Beagle Dog Experimental Model: Safety, immunogenicity, and efficacy. Infect Immun 72: 3252-3259.
- Malfertheiner P, Schultze V, Rosenkranz B, Kaufmann SHE, Ulrichs T, et al. (2008) Safety and immunogenicity of an intramuscular vaccine in noninfected volunteers: A phase I study. Gastroenterology 135: 787-795.
- Monterrubio-Lo´pez GP, Gonza´lez-Y-Merchand JA, Ribas Aparicio RM (2015) Identification of novel potential vaccine candidates against tuberculosis based on reverse vaccinology. Biomed Res Int 2015: 483150.
- Naz A, Awan FM, Obaid A, Muhammad SA, Paracha RZ, et al. (2015) Identification of putative vaccine candidates against Escherichia coli exploiting exoproteome and secretome: A reverse vaccinology-based approach. Infect Genet Evol 32: 280-291.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- International Journal of Diabetes (ISSN: 2644-3031)
- Archive of Obstetrics Gynecology and Reproductive Medicine (ISSN:2640-2297)
- Journal of Infectious Diseases and Research (ISSN: 2688-6537)
- Journal of Pathology and Toxicology Research
- Journal of Rheumatology Research (ISSN:2641-6999)
- International Journal of Internal Medicine and Geriatrics (ISSN: 2689-7687)
- Journal of Carcinogenesis and Mutagenesis Research (ISSN: 2643-0541)









