Research Article
In Silico Surveillance of Multi Drug Resistance Variant Genes of Mycobacterium Tuberculosis
7543
Views & Citations6543
Likes & Shares
The mechanism of drug resistance could portend grave consequences for tuberculosis eradications’ goals of 2050 and presently, it is an impending risk to tuberculosis control programmes in high burden settings. Complete genome nucleotides sequences of M. tuberculosis and their corresponding accession numbers and locations were retrieved, noted, sorted and resolved into Perfect and Strict Resistance genes in CARD database. In Asia “perfect” genes identified were mdsB mutant, mdsA mutant, gols mutant, sdiA mutant, AAC (6')-Iaa mutant, TEM-1 mutant. The mechanism of MDR genes were: Antibiotic efflux: mdsB mutant, mdsA mutant, gols mutant, sdiA mutant while mechanism of antibiotic inactivation was selected for AAC (6')-Iaa mutant, TEM-1 mutant. Perfect MDR variant genes in Europe are: adeK, adeI, abeS, ADC-6, OXA-67 with antibiotic efflux (adeK, adeI, abeS) and antibiotic inactivation (ADC-6, OXA-67). In Africa “perfect” genes identified are efpA mutant, mtrA mutant, gols mutant, mepR mutant, mgrA mutant, norA mutant, arIR mutant, AAC (2')-Ic mutant, BPU-1 mutant, RbpA mutant, mfpA mutant, tetW mutant, mecR1 mutant, mecI mutant. In America “perfect” genes are efpA mutant, mtrA mutant, gols mutant, mepR mutant, mgrA mutant, norA mutant, arIR mutant, mepA mutant, Lmrs mutant, tet(K) mutant, KpnF mutant, oqxA mutant, AAC (2’)-Ic mutant, BPU-1 mutant, FTU-1 mutant, BAT-1 mutant, FosB mutant, ANT (4')-Ib mutant, SHV-1 mutant, mfpA mutant, tetM mutant and ErmA mutant. However, in Antarctica smeD mutant is the only perfect variant gene. Generally, the mechanisms of drug resistance were: efflux, target alteration, target-shielding, target replacement and reduced permeability to antibiotic whereas in Antarctica, Antibiotic efflux, Antibiotic target protection, Antibiotic inactivation, Antibiotic target alteration.
Keywords: CARD-RGI, Multi-drug Resistance, Variant-genes, Whole genome sequences
INTRODUCTION
Tuberculosis is among the leading causes of mortality and fatality worldwide. Drug resistance is becoming a significant world threat to public health. In 2017, the United Nation Agency on infectious disease, reported that there have been associated degree of over 10 million cases of TB and over 3 million deaths reported fatalities throughout the year 2017. Bharat alone accounted for over 24% of world MDR-TB incidence with over twenty seventh of world TB incidence ascertained among HIV-negative people (Global infectious disease UN agency Report [GWTR]x, 2018). A total of 6,520 cases of TB (incidence of roughly twelve per a hundred, were recorded in England, of which 1.4 there have been multidrug-resistant TB (MDR-TB) undiagnosed drug resistance results in higher transmission, leading to poor patient outcomes and likelihood of amplification of drug resistance thereby impeding the preventive strategy of World Health Organization (WHO) to halt TB disease by 2035.
Many studies have shown association of the genetic variations with the pathologic process and drug resistance [2]. The world frontline molecular technology such as the line probe assays and Xpert MTB/RIF used for diagnosing of drug resistant TB are developed on these principles of genetic markers [3]. These have additionally discovered specific genotypes related to drug resistance. Genomics and bioinformatics are more and more contributive to our understanding of infectious diseases caused by microorganisms especially pathogens like mycobacteria. Genomics and bioinformatics have contributed vastly to our understanding of infectious unwellness: from disease pathogenesis, mechanisms and it has also unfolded our understanding of antimicrobial resistance and host’s immune response [4]. Drug-resistant microbes poses a major challenge to infectious disease management programmes in high burden settings. The drug-resistant infectious disease outbreaks in Tugela Falls Ferry and different regions of African nation highlight the requirement for early and correct diagnosing of drug resistance [5]. However, these tests supposed a restricted range of mutations. There are many instances wherever phenotypic resistance could not be explained by known mutations related to drug resistance [6]. A recent study comparison the sensitivity of Xpert MTB/RIF with line probe assay for detection of rifampicin mono-resistant M. tuberculosis according the utility of country specific probes, to increase the sensitivity of Xpert MTB/RIF in Bharat [7]. Since there is wide genetic non-uniformity among M. tuberculosis isolates from totally different geographic regions, large-scale sequencing efforts are urgently needed to map genetic variations and establish the genotypes related to drug resistance.
There were estimated 490,000 new cases of multidrug-resistant (MDR) TB that is outlined by phenotypic resistance to isoniazid and rifampicin [8]. 100% of MDR-TB cases globally will be classified as extensively drug-resistant (XDR), indicating that there's concomitant resistance to quinolones (such as fluoroquinolones, levofloxacin, and moxifloxacin) and to a second-line injectable agent (amikacin, kanamycin, or Capreomycin).
As expected, drug-resistance patterns predict treatment outcome; in 2015, TB treatment success overall was eighty three percent, whereas the success rate was fifty four percent for MDR-TB or rifampicin-resistant TB (RR-TB) and solely half-hour for XDR-TB [9]. Culture-based techniques remained this reference gold standard for diagnosing TB and drug-susceptibility testing of TB, however these processes are long time consuming and need specialized laboratory capability. Additionally, in most recent time, the employment of speedy molecular tests for the diagnosing of TB has enlarged globally, significantly the employment of Xpert MTB/RIF (Cepheid, Sunnyvale, CA), a PCR-based assay that at the same time detects the presence of M. tuberculosis and resistance to rifampicin.
Current recommendations for the treatment of drug-susceptible TB embrace a 6-month course of a multi-drug program of rifampicin, isoniazid, pyrazinamide, and ethambutol. Traditionally, treatment of MDR- or XDR-TB concerned the long-term use of second-line medicine, together with injectable agents (WHO, 2016). Recently, the MDR-TB treatment landscape has modified with the introduction of multiple novels second-line medicine that may be administered orally. In 2012, bedaquiline, a diarylquinolone, became the primary TB drug from a unique drug category to receive North American nation Food and Drug Administration (FDA) approval in over forty years [10]. Another oral agent, delamanid, a nitro-dihydro-imidazooxazole spinoff, has additionally shown promise for TB treatment [11].
WHO revealed updated treatment tips for MDR/RR-TB [12], recommending full oral MDR regimens for several patient groups. The recommended treatment ways embrace each shorter, standardized MDR regimens (for nine to twelve months) and longer, individualized treatment regimens (for eighteen to twenty months). The updated tips cluster opposing tubercular medicine on the idea of however they ought to be combined to form individualized, longer MDR-TB regimens [12].
Despite advances in each medical specialty and medicine for TB, challenges still abound. These obstacles for speedy M. infectious disease diagnosing embrace however not restricted to:
- The imperfect sensitivity of molecular tests for the detection of this infectious agent, significantly within the case of paucibacillary TB (where there's a lower microorganism burden)
- Lack of comprehensive molecular assays because of incomplete information of all resistance mutations in TB
- Technical limitations to the numbers of mutations that may be enclosed on diagnostic molecular platforms.
Furthermore, the rollout of speedy diagnostic platforms to low-resource settings has been a challenge. Remaining treatment challenges include: prolonged treatment courses, resulting in bigger drug exposure, toxicity, and non-compliance; unacceptable side-effect profiles; supplying of drug access; and re-infection [13].
The onset of the new age of genomic sequencing began to revolutionize our approach to human diseases, together with TB. In 1998, [14] the complete genome analysis of the M. tuberculosis reference strain H37Rv, that was just about 4.41 million base pairs long and encoded just about 4000 genes. The primary sequencing of a clinical reference strain, CDC1551, quickly followed [15]. However, even at the time of those breakthroughs, there was recognition that translating these genomic knowledges into clinical profit would prove difficult. Despite these challenges, it's clear, over twenty years later, that M. tuberculosis genomic data are remarkably helpful in raising our understanding of how drug-resistant TB evolves and spreads and in serving to inform medical diagnosis and therapies [16].
Comprehensive Antibiotic Resistance Database is a biological database that collects and organizes reference information on antimicrobial resistance genes, proteins and phenotypes [17]. The database covers all types of drug classes and resistance mechanisms and structures its data based on ontology. The CARD database was one of the first resources that covered antimicrobial resistance genes [18].
MATERIALS AND METHODS
Retrieval of complete genome sequence of Mycobacterium tuberculosis
In Silico AMR detection was performed by using a search query input of nucleotides sequences for the presence of pre-determined set of AMR determinants as contained in CARD databases. A total of 85 different complete genome sequences in FASTA format of Mycobacterium tuberculosis were retrieved from NCBI nucleotide database on 19th July, 2020.
Detection of antibiotic Resistant genes in Mycobacterium tuberculosis
The complete genome sequences of Mycobacterium tuberculosis were analyzed to detect the presence or absence of antibiotic resistant genes and mutants Analysis were carried out using the Comprehensive Antibiotic Resistance Database (CARD). The Resistant Gene Identifier (RGI) was employed for detection of the resistant genes and mutants present. The AMR genes were categorized as perfect and strict. The RGI charts were downloaded from the CARD databases into excel datasheet for chart analysis (Figure 1).
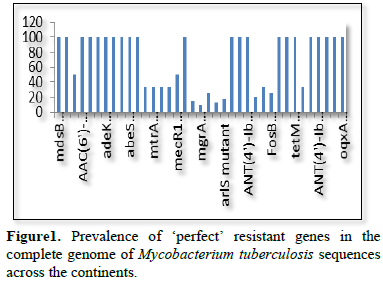

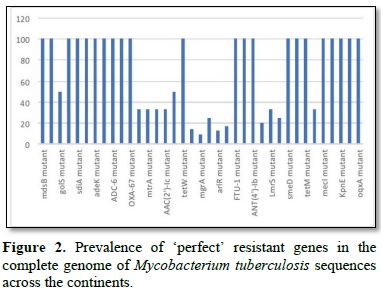
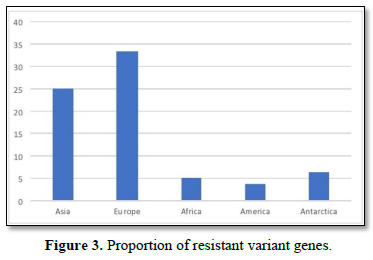
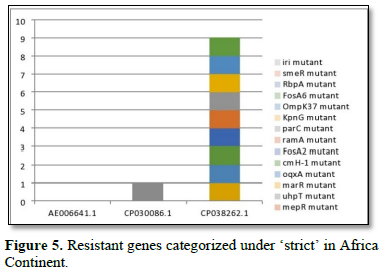
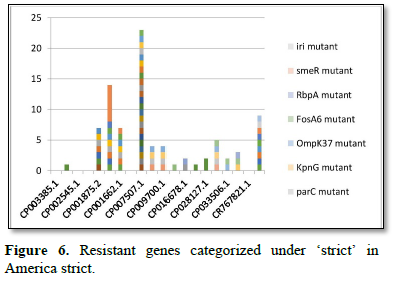
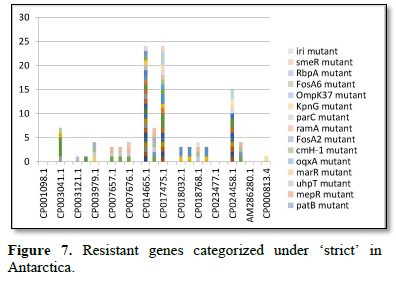
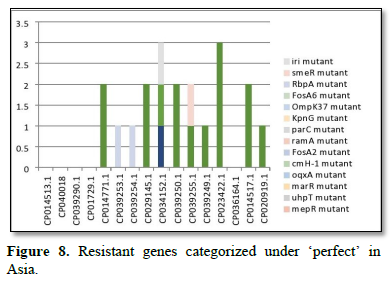
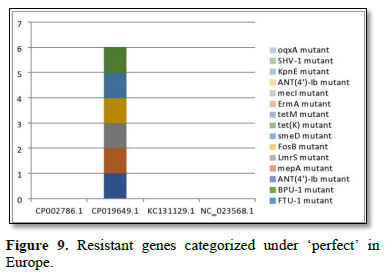
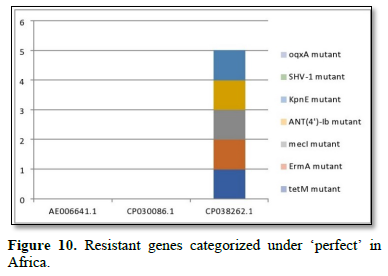
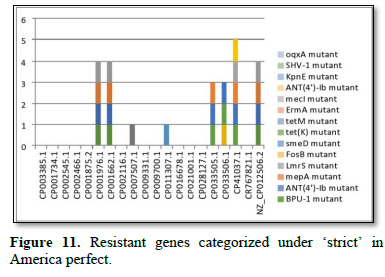
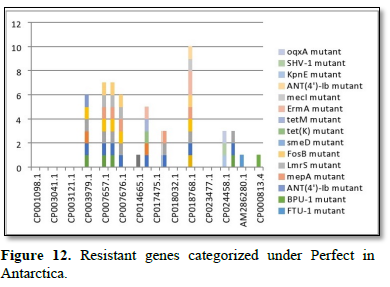
Figure 2 shows the prevalence of resistant of genes categorized under ‘perfect’ as obtained after analysis using the CARD (Comprehensive Antibiotic Resistance Database) resistant genes identifier. mdsB, mdsA, AAC (6')-Iaa, sdiA, TEM-1, adeK, adeI, ADC-6, abeS, OXA-67, tetW, FTU-1, BPU-1, ANT (4')-Ib, smeD, tet(K), tetM, mecI, ANT (4')-Ib, KpnE, SHV-1, oqxA had80% prevalence each (present in all the 85 complete genome sequences retrieved), golS and mecR1 had 47% prevalence, efpA, mtrA, mfpA, AAC (2')-Ic, LmrS, ErmA had 26.3% prevalence each, norA and FosB had 24.3% prevalence each. arlS, mepR, mgrA had the prevalence rates of 15.72%, 16.72%, and 16.95% respectively. The presence of genes under the ‘perfect’ category from CARD-RGI analysis in each of the whole genome sequences represented by their accession numbers is shown in Figure 3. Percentage distribution of ‘strict’ resistant genes present in the complete genome of Mycobacterium tuberculosis sequences. In the same vein Figure 4 shows the prevalence of genes categorized under the strict category from CARD database with APH (6)-Id, APH (3'')-Ib, sul2, tet(B), ANT (3'')-Iic, adeJ, adeL, AmvA, adeN, adeR, AbaF, KpnH, gyrB, bacA, vanI, farB, mecR1, tet (45), cat86, rpsL, pncA, soxR, patB, oqxA, cmH-1, FosA2, ramA, parC, KpnG, OmpK37, FosA6, smeR, iri(100%) each. mdfA, AbaQ, gyrA, thyA, kasA, AAC (6')-Iy, emrB, Bla2, Bla1, mphL, arlR, blaZ, RbpA, mepR, marR (50%) each. folC, acrB, acrA, kdpE, KpnF, MdtK, folP, adeF. EF-Tu, RbpA, rpoB, katG, sdiA, GlpT, uhpT (33.33%) each. PmrF, ampC1, CRP, embB, FosB (25%) each. msbA and emrR had (20%) each. ampH, KpnE, marA, H-NS, baeR (16%) each. mecA and norA (14%) each. murA, rRNA and LmrS (12%) each. The presence of genes under the ‘strict’ category from CARD-RGI analysis in each of the whole genome sequences represented by their accession number is shown in Figure 5, the prevalence of ‘perfect’ resistant genes presents in the complete genome of Mycobacterium tuberculosis sequences and their clinical implication in five (5) continents (Figures 6-12).











DISCUSSION
Comprehensive Antibiotic Resistance Database resistant genes identifier (CARD-RGI) have been successfully used in this study to understand antibiotic resistance genes in about 85 complete genome sequences and accessions of Mycobacterium tuberculosis retrieved from the NCBI. We retrieved 85 complete genomes from the NCBI database of Mycobacterium tuberculosis with the lowest number of complete genome sequences retrieved from Asia and Europe, (3% and 4% respectively) due to the limited number of the complete genome sequences of Mycobacterium tuberculosis deposited in the NCBI GenBank from these locations. Drug resistance in M. tuberculosis often arises as a result of DNA chromosomal aberrations in existing genes that are of vertical descent. Lateral genetic exchanges are rare in M. tuberculosis because they lack plasmid [19]. The idea of strict, perfect and loose genes suggested an indication of the degree of variant MDR genes in bacteria [20].
The resistance variant genes in this study are divided into perfect and strict. In all as expected, we resolved less of ‘perfects’ genes than ‘stricts’. These are: APH(6)-Id mutant, APH(3'')-Ib mutant, sul2 mutant, tet(B) mutant, ANT(3'')-Iic mutant, adeJ mutant, adeL mutant, AmvA mutant, adeN mutant, adeR mutant, AbaF mutant, KpnH mutant, gyrB mutant, bacA mutant, vanI mutant, farB mutant, mecR1 mutant, tet(45) mutant ,cat86 mutant, rpsL mutant, pncA mutant, soxRv mutant, patB mutant, oqxA mutant, cmH-1 mutant, FosA2 mutant, ramA mutant, parC mutant, KpnG mutant, OmpK37 mutant, FosA6 mutant, smeR mutant, iri mutant, mdfA mutant, AbaQ mutant, gyrA mutant, thyA mutant, kasA mutant, AAC(6')-Iy mutant, emrB mutant, Bla2 mutant, Bla1 mutant, mphL mutant, arlR mutant, blaZ mutant, RbpA mutant, mepR mutant, marR mutant, folC mutant, acrB mutant, acrA mutant, kdpE mutant, KpnF mutant, MdtK mutant, folP mutant, adeF mutant, EF-Tu mutant, RbpA mutant, rpoB mutant, katG mutant, sdiA mutant, GlpT mutant, uhpT mutant, PmrF mutant, ampC1 mutant, CRP mutant embB mutant, FosB mutant, msbA mutant, emrR mutant, ampH mutant, KpnE mutant, marA mutant, H-NS mutant, baeR mutant, mecA mutant, norA mutant, murA mutant, rRNA mutant, LmrS mutant, AAC(6')-Iaa mutant, mtrA mutant, AAC(2')-Ic mutant, SHV-1 mutant, efpA mutant, tetM mutant, tet(K) mutant, FTU-1 mutant, BPU-1 mutant, ANT(4')-Ib mutant, smeD mutant, mecI mutant, ADC-6 mutant, abeS mutant OXA-67 mutant, mdsB mutant, mdsA mutant, TEM-1 mutant, adeK mutant, adeI mutant, tetW mutant , ANT(4')-Ib mutant, golS mutant, mecR1 mutant, ErmA mutant, mfpA mutant, have been successfully identified. The drug-resistant tuberculosis outbreaks in Tugela Ferry and other regions of South Africa highlight the need for early and accurate diagnosis of drug resistance [5]. There are reported studies of established association of the genetic variations with pathogenesis and drug resistance [21]. Global frontline molecular diagnostics such as line probe assays and Xpert MTB/RIF used for diagnosis of drug resistant TB have been developed based on these genetic markers [3]. This study corroborated our earlier study on a similar subject which re-established phenotypic resistance could not be explained by known mutations associated with drug resistance [6]. Based on this mechanism a recent study compared the efficacy of Xpert MTB/RIF with line probe assay for detection of rifampicin mono-resistant M. tuberculosis [7]. This study noticed a considerable genetic heterogeneity among M. tuberculosis isolates across the continents, large-scale sequencing efforts to map genetic variations and identify the genotypes associated with drug resistance. Prevalence of ‘perfect’ resistant genes present in the complete genome of Mycobacterium tuberculosis sequences and their clinical implication in five (5) continents: In Asia “perfect” Antibiotic efflux (mdsB mutant, mdsA mutant, gols mutant, sdiA mutant), Antibiotic inactivation (AAC (6')-Iaa mutant, TEM-1 mutant). In Europe “perfect” Antibiotic efflux (adeK, adeI, abeS), Antibiotic inactivation (ADC-6, OXA-67). In Africa “perfect” Antibiotic efflux (efpA mutant, mtrA mutant, gols mutant, mepR mutant, mgrA mutant, norA mutant, arIR mutant), Antibiotic inactivation (AAC (2')-Ic mutant, BPU-1 mutant), Antibiotic target protection (RbpA mutant, mfpA mutant, tetW mutant) Antibiotic target replacement (mecR1 mutant, mecI mutant). In America “perfect” Antibiotics efflux (efpA mutant, mtrA mutant, gols mutant, mepR mutant, mgrA mutant, norA mutant, arIR mutant, mepA mutant, Lmrs mutant, tet(K) mutant, KpnF mutant, oqxA mutant), Antibiotic inactivation (AAC (2’)-Ic mutant, BPU-1 mutant, FTU-1 mutant, BAT-1 mutant, FosB mutant, ANT(4')-Ib mutant, SHV-1 mutant), Antibiotic target protection (mfpA mutant, tetM mutant), Antibiotic target alteration (ErmA mutant). In Antarctica “perfect” Antibiotic efflux (smeD mutant). Prevalence of ‘strict’ resistant genes present in the complete genome of Mycobacterium tuberculosis sequences and their clinical implication in five (5) continents: In Asia “strict” Antibiotic efflux (acrB mutant, acrA mutant, kdpE mutant, mdfA mutant, msbA mutant, kpnE mutant, H-NS mutant, matK mutant, baeR mutant, tet(B) mutant, emrR mutant), Antibiotic inactivation (ampH mutant, ampC1 mutant, APH(6)-Id mutant, APH(3'')-Ib mutant), Antibiotic target alteration (folC mutant, pmrF mutant, sul2 mutant), Antibiotic efflux, reduced permeability to antibiotic (MarA mutant). In Europe “strict”Antibiotic efflux (adeJ mutant, adeF mutant, adeL mutant, AmvA mutant, adeN mutant, AbaQ mutant, adeR mutant, AbaF mutant), Antibiotic inactivation (ANT(3'')-Iic mutant), Antibiotic target alteration (folP mutant, parC mutant). In Africa “strict” Antibiotic efflux (acrB mutant, acrA mutant, KdpE mutant, mdfA mutant, msbA mutant, KpnF mutant, H-NS mutant, baeR mutant, tet(45) mutant, emrR mutant, adeF mutant, emrB mutant, sdiA mutant, FarB mutant, CRP mutant, KnpH mutant, arIR mutant, Lmrs mutant, norA mutant), Antibiotic inactivation (ampH mutant, ampC1 mutant, Bla2 mutant, Bla1 mutant, FosB mutant, mphL mutant, PC1 mutant, AAC(6')-Iy mutant), Antibiotic target alteration (gyrB mutant, PBP3 mutation, EF-Tu mutation, gyrA mutation, thyA mutation, KasA mutation, Kat mutation, murA mutation, embB mutation, rRNA mutation, KatG mutation, pmrF mutation, bacA mutation, GlpT mutation, VanI mutation, FolP mutation, rpsL mutation, pncA mutation), Antibiotic target alteration, antibiotic target replacement (rpoB mutation), Antibiotic target protection (RbpA mutation, adeF mutation), Antibiotic efflux, reduced permeability to antibiotic (marA mutation), Antibiotic target replacement (mecR1 mutation, mecA mutation).
In America “strict”Antibiotic efflux (norA mutation, acrB mutant, acrA mutant, KdpE mutant, mdfA mutant, msbA mutant, KpnF mutant, H-NS mutant, baeR mutant, tet(45) mutant, emrR mutant, adeF mutant, emrB mutant, sdiA mutant, FarB mutant, CRP mutant, KnpE mutant, arIR mutant, Lmrs mutant, AbaQ mutation, patB mutation, mepR mutation, mdtK mutation, mdfA mutation, kdpDE mutation, oqxA mutation, KpnG mutation, KpnH mutation), Antibiotic target alteration (GlpT mutation, murA mutation, gyr mutation, KatG mutation, pncA mutation, embB mutation, rRNA mutation, rspL mutation, FoIP mutation, norA mutation, baeA mutation, PmrF mutation, UhpT mutation, EF-Tu mutation, PBP3 mutation, parC mutation), Antibiotic inactivation (Cat86 mutation, blaz mutation, FosB mutation, ampC1 mutation, ampH mutation, CMH-1 mutation, FosA2 mutation, FosA6 mutation), Antibiotic target protection (RbpA mutation), Antibiotic target alteration, antibiotic target replacement (rpoB mutation), Antibiotic target replacement (mecA mutation), Antibiotic efflux, reduced permeability to antibiotic (MarA mutation, ramA mutation, ompK37 mutation). In Antarctica “strict” Antibiotic efflux (adeF mutation, smeR mutation), Antibiotic target protection (RbpA mutation), Antibiotic inactivation (iri mutation), Antibiotic target alteration (folC mutation). Spontaneous chromosomal mutations are the genetic basis for drug resistance in M. tuberculosis [21], and a limited number of mutations account for the majority of phenotypic resistance to anti-TB drugs. RIF interacts with the ß-subunit (encoded by rpoB) of the RNA polymerase and inhibits the early steps of transcription [22]. Resistance to RIF is almost entirely coupled to mutations within an 81-bp region of the rpoB gene, called the RIF resistance-determining region (RRDR), comprising codons 507 to 533 [23].
A mutation at nucleotide 1401 or 1484 is associated with resistance to all these agents, whereas a mutation at nucleotide 1402 is associated with CAP resistance and low-level KAN resistance [24]. Overexpression of eis (encoding the aminoglycoside acetyltransferase Eis), caused by mutations in the promoter region, confers low-level resistance to KAN [25] Fluoroquinolones (FQs), e.g., Ofloxacin (OFL), which bind to DNA gyrase, inhibit proper regulation of supercoiling and cause chromosomal double-strand breaks [26]. DNA gyrase is a hetero tetramer consisting of two A and two B subunits, encoded by the genes gyrA and gyrB [27]. Since sequences are highly conserved, certain mutations correlate well with phenotypic resistance, and a limited number of mutations account for the majority of phenotypic resistance to the important anti-tuberculosis medications, various methods of genotypic testing have successfully been used for the rapid detection of M. tuberculosis resistance [28].
CONCLUSION
Earlier studies revealed that as much as drug-resistance are induced and acquired; mixed infections are common, and often represent important intermediates in the evolution of drug resistance. Furthermore, its practical use in clinical settings holds great potential to improve public health through real-time molecular epidemiology tracking, to identify global hotspots of drug-resistance emergence, and to facilitate the development of improved approaches for the diagnosis and treatment of drug-resistant Mycobacterium tuberculosis.
DECLARATION OF CONFLICTING INTERESTS
The authors declared no conflict of interests whatsoever.
- Gupta R, Lavollay M, Mainardi JL, Arthur M, Bishai WR, et al. (2010) The Mycobacterium tuberculosis protein LdtMt2 is a nonclassical transpeptidase required for virulence and resistance to amoxicillin. Nat Med 16: 466-469.
- Laurenzo D, Mousa SA (2011) Mechanisms of drug resistance in Mycobacterium tuberculosis and current status of rapid molecular diagnostic testing. Acta Trop 119(1): 5-10.
- Gagneux S, Small PM (2007) Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7(5): 328-337.
- Long S, Beres S, Olsen R, Musser J (2014) Absence of patient-to-patient intra hospital transmission of Staphylococcus aureus as determined by whole-genome sequencing. mBio 5(5): e01692-e1714.
- Casali N, Nikolayevskyy V, Balabanova Y, Harris SR, Ignatyeva O, et al. (2014) (2014) Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet 46(3): 279-286.
- Rigouts L, Gumusboga M, De Rijk WB, Nduwamahoro E, Uwizeye C, et al. (2013) Rifampin resistance missed in automated liquid culture system for Mycobacterium tuberculosis isolates with specific rpoB mutations. J Clin Microbiol 51(8): 2641-2645.
- Rufai SB, Kumar P, Singh A, Prajapati S, Balooni V, et al. (2014) Comparison of Xpert MTB/RIF with line probe assay for detection of rifampin-mono resistant Mycobacterium tuberculosis. J Clin Microbiol 52(6): 1846-1852.
- World Health Organization (2016) WHO treatment guidelines for drug-resistant tuberculosis. 2016 update. Geneva. Available online at: https://www.who.int/publications/i/item/9789241549639
- World Health Organization (2017) TB burden estimates, notifications and treatment outcomes for individual countries and territories, who regions and the world. Available online at: https://www.who.int/tb/publications/global_report/gtbr2017_annex4.pdf
- Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, et al. (2012) Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: Long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother 56:3271-3276.
- Matsumoto M, Hashizume H, Tomishige T, Kawasaki M, Tsubouchi H, et al. (2006) OPC-67683, a nitro-dihydro-imidazooxazole derivative with promising action against tuberculosis in vitro and in mice. PLoS Med 3: 2131-2144.
- World Health Organization (2018) Global Tuberculosis Report 2018. Geneva. Available online at: https://apps.who.int/iris/bitstream/handle/10665/274453/9789241565646-eng.pdf?sequence=1&isAllowed=y
- Bloom BR, Atun R, Cohen T, Dye C, Fraser H, et al. (2017) Tuberculosis. In: Holmes KK, editors. Major infectious diseases. Washington DC: The International Bank for Reconstruction and Development/The World Bank. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK525174/
- Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, et al. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393: 537-544.
- Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, et al. (2002) Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol 184: 5479-5490.
- Cabibbe AM, Walker TM, Niemann S, Cirillo DM (2018) Whole genome sequencing of Mycobacterium tuberculosis. Eur Respir J 52: 537-545.
- Jia B, Raphenya AR, Alcock B, Waglechner N, Guo P, et al. (2017) CARD 2017: Expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res 45(D1): D566-D573.
- Arthur Y, Ehebauer MT, Mukhopadhyay S, Hasnain SE (2013) The PE/PPE multi gene family codes for virulence factors and is a possible source of mycobacterial antigenic variation: Perhaps more? Biochimie 94: 110-116.
- Dubée V, Triboulet S, Mainardi J, Ethève-Quelquejeu M, Marie A, et al. (2012) Inactivation of Mycobacterium tuberculosis l, d-transpeptidase LdtMt1 by carbapenems and cephalosporins. Antimicrob Agents Chemother 56: 4189-4195.
- Alcock BP, Raphenya AR, Lau TTY, Tsang KK, Bouchard M, et al. (2019) CARD 2020: Antibiotic resistomes surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48(D1): D517-D525.
- Zhang Y, Yew WW (2009) Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 13: 1320-1330.
- Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, et al. (2001) Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell 104: 901-912.
- Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, et al. (2009) Tuberculosis drug resistance mutation database. PLoS Med 6(2): e2.
- Engström A, Perskvist N, Werngren J, Hoffner SE, Jureen P (2011) Comparison of clinical isolates and in vitro selected mutants reveals that tlyA is not a sensitive genetic marker for capreomycin resistance in Mycobacterium tuberculosis. J Antimicrob Chemother 66: 1247-1254.
- Zaunbrecher MA, Sikes RD Jr, Metchock B, Shinnick TM, Posey JE (2009) Overexpression of the chromosomally encoded aminoglycoside acetyltransferase eis confers kanamycin resistance in Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 106: 20004-20009.
- Xu C, Kreiswirth BN, Sreevatsan S, Musser JM, Drlica K (1996) Fluoroquinolone resistance associated with specific gyrase mutations in clinical isolates of multidrug-resistant Mycobacterium tuberculosis. J Infect Dis 174: 1127-1130.
- Piton J, Petrella S, Delarue M, André-Leroux G, Jarlier V, et al. (2010) Structural insights into the quinolone resistance mechanism of Mycobacterium tuberculosis DNA gyrase. PloS One 5(8): e12245.
- Palomino JC (2006) Newer diagnostics for tuberculosis and multi-drug resistant tuberculosis. Curr Opin Pulm Med 12: 172-178.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Journal of Otolaryngology and Neurotology Research(ISSN:2641-6956)
- Journal of Pathology and Toxicology Research
- Chemotherapy Research Journal (ISSN:2642-0236)
- International Journal of Internal Medicine and Geriatrics (ISSN: 2689-7687)
- Journal of Psychiatry and Psychology Research (ISSN:2640-6136)
- Archive of Obstetrics Gynecology and Reproductive Medicine (ISSN:2640-2297)
- Journal of Rheumatology Research (ISSN:2641-6999)














