Research Article
Molecular Surveillance of Regional S. Pneumoniae Serotype Distribution in Invasive Pneumococcal Disease Among Children Across India
5869
Views & Citations4869
Likes & Shares
Background: Serotype distribution of pneumococci varies with age, time and geographical area. Limited data is available on region specific serotype prevalence of Pneumococci in India. This study reports the country specific prevalence data generated using PCRSeqTyping. This molecular typing strategy was improvised and standardized at our center to type 91 serotypes uniquely from serum samples.
Methods: 1504 serum samples were collected across 7 regions of India (Delhi-234, Kanpur-200, Kolkata42, Ludhiana-87, Jodhpur-234, Bangalore-351 and Chennai-250) from children below 5 years with clinically suspected, radiological confirmed Pneumonia in addition to raised CBC, CRP and Procalcitonin. These samples were subjected to real time multiplex PCR for S. pneumoniae identification. 456 samples tested positive for S. pneumoniae were further subjected to PCRSeqTyping for serotype detection.
Results: The qmPCR positivity across the regions ranged from 28%- 32%. The 456 qmPCR positive samples belonged to 30 different serotypes among which 17 were Non-vaccine types. The percentage prevalence of the common serotypes 1, 6B, 14,19F & 23F differed regionally. Serotypes 9V, 3&4, 7F, 9N & 23F were unique to Delhi, Kanpur, Jodhpur, Bangalore and Chennai regions correspondingly. Serotype-1 isolation was maximum at Delhi and least at Chennai.
Conclusions: The study emphasizes the need of a comprehensive and continued region wise reporting on serotypes/serogroups. A robust understanding of the serotypes in different regions, as well as linkage of data at national level would help produce wide ranging inputs. It is of importance from an epidemiological and public health perspective for the development of vaccines and policies. The added advantage of sensitive, specific, economical molecular typing methodology for pneumococcal surveillance is presented.
Keywords: S. pneumoniae, PCRSeqTyping, Culture negative, qmPCR, Serotyping
Abbreviations
IPD: Invasive Pneumococcal Disease, qmPCR: Quantitative multiplex real-time PCR, DNA: Deoxy Ribonucleic Acid, PCR: Polymerase chain reaction, WHO: World Health Organization, CBC: Complete Blood Count, CRP: C-Reaction Protein, PCT: Procalcitonin, GAPDh: Glyceraldehyde 3-phosphate dehydrogenase, PCV: Pneumococcal Conjugate Vaccine
INTRODUCTION
Since 1990, both the incidence and the number of infant deaths has been decreased by more than half worldwide (from 93 deaths per 1,000 live births in 1990 to an estimated 38 in 2019). Despite the global progress in reducing child mortality over the past few decades, an estimated 5.2 million children under age five died in 2019-Nearly half (49 per cent) of all under-five deaths in 2019 occurred in five countries: Nigeria, India, Pakistan, the Democratic Republic of the Congo and Ethiopia. Nigeria and India alone account for almost a third. This highlights that as we look past the MDGs, child survival needs to be an ongoing focus. In achieving the under-5 mortality rate targets, India has been lagging behind. India is one of the world's top countries with the largest number of child deaths [1].
Understanding the causes of infant mortality offers valuable insights into public health. About half of the global under-five deaths are caused by infectious diseases (such as pneumonia and diarrhea). Pneumonia accounts for 16% of under-five deaths, with 55.8% of the death fraction due pneumococcal pneumonia being observed [2]. In Indian research, the incidence of invasive pneumococcal disease (IPD) in children admitted to hospitals with suspected invasive bacterial diseases was 10.58 percent, and 24 percent of all cases of bacterial pneumonia were due to S. pneumoniae [3]. It remains difficult to estimate the exact burden of pneumococcal disease in India, though morbidity and mortality rates remain high.
- pneumoniae vaccine probe studies have shown that the actual incidence of pneumococcal pneumonia is much higher than initially suspected and the methods of blood culture to detect S. pneumoniae catch just approximately 10-20 percent of suspected pneumococcal pneumonia cases, with the sensitivity in infants being even lower [4].
Studies on pneumococcal disease and its epidemiology are compounded by problems associated with establishing an etiological diagnosis to determine serotype prevalence. Improved methods for diagnosing pneumonia are needed to better inform health policy-makers. Information on disease burden and circulating serotypes can build local awareness of disease, enable evaluation of health impacts resulting from interventions, and inform treatment priorities and prevention strategies. Molecular detection methods like Quantitative multiplex real-time PCR (qmPCR) have facilitated an improvement in pneumococcal disease diagnosis, as it offers a swift, more sensitive and specific strategy.
Currently available tests for pneumococcal serotyping including conventional Quellung test and DNA-based tests, have limitations and variable performance in different settings. For many developing countries, it is crucial to find a robust method with complete serotype coverage which is practical, simple and cost-effective for routine serotype prediction and pneumococcal serogroup/serotype surveillance [5]. Herein, we apply an innovative serotyping approach, using the dynamics of sequencing technology, in a 2-step sequencing-based typing system for serotyping pneumococci directly from serum samples [6]. This method relies on sequencing of assembly genes located in the capsular operon to identify all pneumococcal serotypes.
The aim of this study was to apply qmPCR followed by sequencing-based technology for rapid identification and serotyping of pneumococci directly from culture negative clinical samples which would give the true estimate of disease burden and circulating serotypes prevalent in India [7].
MATERIALS AND METHODS
Study population
1504 pediatric patients aged 28 days to 60 months were enrolled for the study. The inclusion criteria for the patients involved were, pneumonia meeting WHO clinical criteria. Patients with history of measured temperature of ≥38°C within 24 hours before screening with suspected bacterial infection due to S. pneumoniae or other invasive pneumococcal disease with raised CBC, CRP and positive PCT test were also included in the study.
Sample collection
Venous blood sample (5ml) was collected from each child; 3ml of blood was directly inserted into blood culture bottle, incubated at 37ºC and 2ml was immediately transferred to sterile vacutainer tube for serum separation.
DNA extraction for qmPCR analysis and PCRSeqTyping
Previously frozen (-80°C) serum specimens were subjected to DNA extraction using QIAamp DNA Mini Kit with automated DNA extractor, QIA cube (Qiagen, Germany), as per manufacturer’s protocol. Quantity and quality of the extracted DNA was determined spectrophotometrically at absorbance 260 nm using Nanodrop 2000 (Thermo Fisher Scientific, USA).
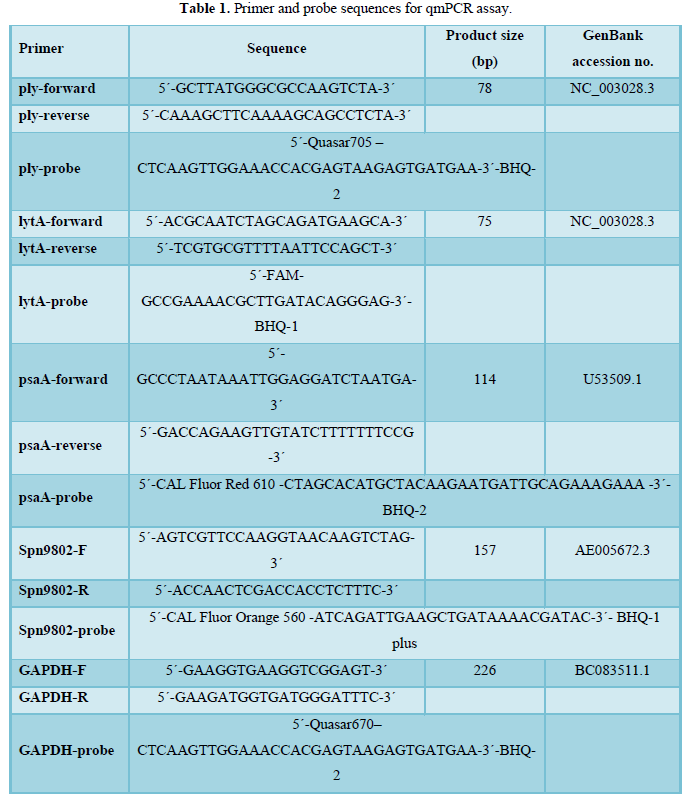
qmPCR assay for detection of S. pneumoniae
The qmPCR assay was performed immediately after DNA extraction. The oligonucleotide 140 sequences for ply, lytA, psaA, Spn9802, and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) 141 primer-probe sets were selected and synthesized according to our earlier published data [7]. The 142 primer and probe sequences are listed in Table 1. qmPCR assay was carried out as described in 143 detail in our previous publication [7]. Briefly, 20X primer-probe mix comprising 10 μM of each 144-forward primer, 10 μM of each reverse primer, and 4 μM of each hydrolysis probe was prepared 145 in DNase/RNase free water (Qiagen) from 100 μM stock solutions of each primer and hydrolysis 146 probe. The real-time PCR amplifications were performed in 25-μl reaction volumes containing 2X 147 Rotor-Gene Multiplex PCR Master Mix (Qiagen), 20X primer-probe mix, template DNA (5 ul) 148 and water. All reactions were performed in duplicate, and for amplification and detection, Rotor149 Gene® Q (Qiagen) was used. Standard amplification parameters were used and were as follows: 150 1 cycle of denaturation at 95°C for 5 min, followed by 40 cycles of denaturation at 95°C for 30 s 151 and combined annealing/extension step at 60°C for 15 s. Data analysis was performed by using 152 Rotor-Gene® Q software (Qiagen).
Molecular Serotyping of Serum samples
qmPCR positive samples were subjected to molecular serotyping using PCRSeqTyping protocol 159 [6]. PCRSeqTyping assay was performed in 2-step. First step involved PCR amplification and 160 sequencing of cpsB gene from genomic DNA of pneumococcal isolates and serum samples. The 161 serotypes were grouped into non-homologous (Group I) and homologous (Group II). Homologous 162 group was further subdivided into ten subgroups and were differentiated based on group specific 163 primers in second step of PCR and sequencing.

PCR amplification
PCR reaction was performed using the primers designed by Leung [8], with modifications. Primers used in the study were cps1-FP(5’-GCAATGCCAGACAGTAACCTCTAT3)’, cps2RP(5’CCTGCCTGCAAGTCTTGATT-3’) and cps-2538-RP (5’-CTTTACCAACCTTTGTAATCCAT-3’). The reaction mixture was modified to contain 50 – 100 ng of genomic DNA, 0.75 Units XT-5 polymerase (3 U/l) (Merck), 1X XT5A-Assay buffer (10X), 1 ul deoxy nucleoside triphosphates (dNTPs, 2.5 mM each, Fermentas), and 100 ng each primer, made up to a final volume of 25 l with DNase/RNase-free distilled water (Gibco). Thermal cycling was performed in the Gene Amp PCR system 9700 (Applied Biosystems) under the following conditions: 94°C for 5 min, followed by 35 amplification cycles of 94°C for 30 s, 50°C for 30 s, and 72°C for 1 min, then a final extension at 72 C for 5 mins. The PCR products were separated by electrophoresis on 1.2 % agarose gel for 45 min at 80 V in 1X Tris-acetate EDTA buffer. Ethidium bromide-stained DNA products were visualized under UV illumination and sized by using a 1–kb DNA molecular size marker (Fermentas).
Sequencing and data analysis
PCR products were subjected to cycle sequencing employing the Big dye Sequence Terminator kit V3.1 (Applied Biosystems) and analyzed on an ABI 3730 XL Genetic Analyzer (Applied Biosystems). Sequencing was performed in one direction using forward primer (cps1), 5’-GCA ATG CCA GAC AGT AAC CTC TAT-3’ by using Long Seq Module (ABI). DNA sequences obtained were used to interrogate the GenBank database (http://www.ncbi.nlm.nih.gov/blast/) and assign serotype using the criteria as per protocol [9]. Serotype of the cpsB nucleotide sequence from GenBank with the highest BLAST bit score was assigned, provided that sequence identity was >99% with the query amplicon nucleotide sequence.
Homology group assignment and PCRSeqTyping
For the homologous strain, a second round of PCR was performed using the primers as specified in the Table 2. PCR products were subjected to cycle sequencing reaction. The nucleotide sequence data was used to assign the serotype.

RESULTS
Demographic and clinical characteristics
Out of 1504 patients, 494 were from 2-5 years of age and 1010 were
qmPCR results from serum samples
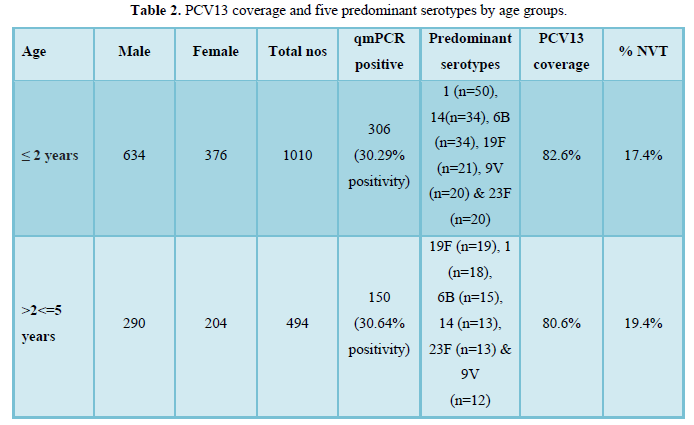
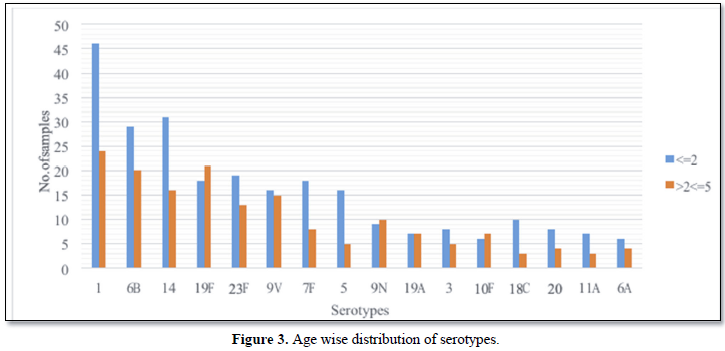
Serum qmPCR for S. pneumoniae was positive in 456 (30%) of 1504 patients. 456 serum samples were from children <=2 years age group (67.1%, n=306) and >2<=5 years age group (32.9%, n=150).
Molecular serotyping of culture negative serum samples
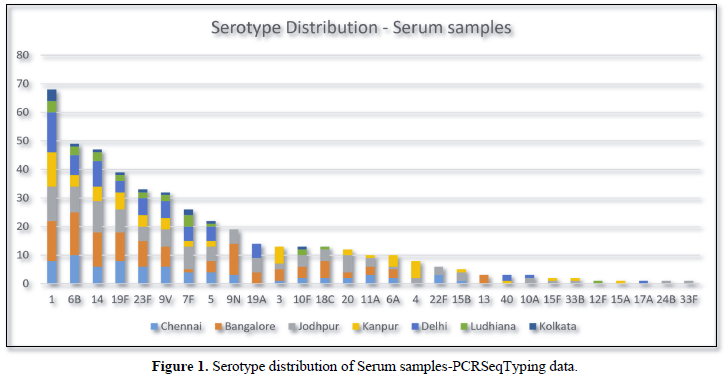
PCRSeqTyping, performed on qmPCR positive clinical specimens i.e., Serum (n=456), identified serotype in all the samples up to type level. Four hundred and fifty-six samples were distributed among 29 different serotypes (Figure 1). Serotypes 1, 6B, 14, 19F and 23F were predominant, covering up to 51.9% (n=237/456). Vaccine coverage by PCV7, PCV10 and PCV13 was found to be 48%, 73.7% and 81.8% respectively. Non vaccine serotypes were observed in 18.2% of the samples (n=83). Among the non-vaccine serotypes, serotypes 9N (n=19), 10F (n=13), 20 (n=12), 11A (n=10) and 22F (n=6) were predominant.
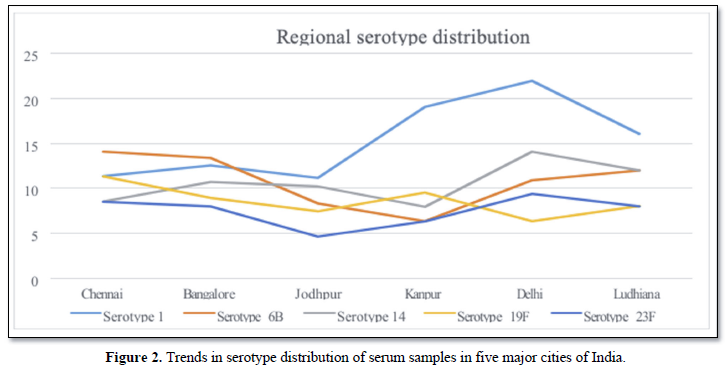
Figure 2 shows the regional distribution of serotypes from five different cities, Chennai, Bangalore, Jodhpur, Kanpur and Delhi.
67.1% (n=306/456) of the serum samples were derived from children 2 to (Figure 3) [3].



DISCUSSION
Isolation of S. pneumoniae from a normally sterile body site provides conclusive evidence of invasive pneumococcal infection, but this is achieved only for a minority of cases [10]. Despite the global importance of pneumococcal disease, there have been surprisingly few recent developments in laboratory diagnostics. [10,11] The capacity to obtain accurate data on IPD burden and to assess the effectiveness of vaccination is hindered by the limitations of existing diagnostic tests. To conquer these limitations, we developed and evaluated a highly sensitive and specific quantitative multiplex real time PCR assay for the identification of S. pneumoniae from culture positive and culture negative samples.
Several molecular PCR typing protocols for S. pneumoniae are described targeting serotype specific regions of cps. [9,12-16]. Nevertheless, these methods were of low resolution between cross reactive serotypes and differentiation of cross-reactive serotypes required a high number of primers. Methods for rapid and reliable serotyping of Streptococcus pneumoniae not based on bacterial culture are needed in the epidemiological surveillance of pneumococcal infection. The development of PCR-based serotyping assays performed directly on clinical samples that are not required to contain viable bacteria has the potential to overcome some of the difficulties associated with these microbiological diagnostic procedures that can perform poorly, especially in pediatric patients [16]. We have developed a PCR and sequencing-based assay to identify all known and emerging serotypes from clinical isolates. In the present study, we report the performance of qmPCR and PCRSeqTyping assay directly on clinical samples from culture negative patients.
Serotype distribution of the pneumococcus changes with time due to various factors including clonal enlargement, capsular transformation, mass and routine pneumococcal vaccination, and widespread antibiotic use in population. Pneumococcal serotype appears to be much more important in determining colonization, disease development, and clinical phenotype compared to the genetic background [17]. Thus, obtaining information about local serotype distribution of pneumococci and observing potential changes in the distribution in time is essential for effective vaccination strategies. The most common serotypes causing IPD in European children are 14, 6B, 19F and 23F, which are included in PCV7 [18]. A recent global review on pneumococcal serotypes causing invasive disease in 51 countries showed that serotype 14 was the most common in all regions including Africa, Asia, Europe, Latin America, North America and Oceania [19]. They observed that seven serotypes 1, 5, 6A, 6B, 14, 19F and 23F accounted for 66-76% of diseases in each region, and the top eight serotypes in Africa and Asia were the same which included, the seven serotypes mentioned in addition to serotype 19A. It was also observed that serotype 14 was the dominant serotype in younger children (
Estimating the true pneumococcal disease burden in Indian continues to be challenging, while morbidity and mortality rates remain high [20]. Hence, studies on serotype prevalence are still relevant in the Indian context. There is a paucity in data on serotypes from India as some of the studies conducted had limited sample size and failed to contribute to epidemiological analysis [21]. Some of the studies were conducted almost a decade back during which the epidemiology of S. pneumoniae might have changed to a great extent [22].
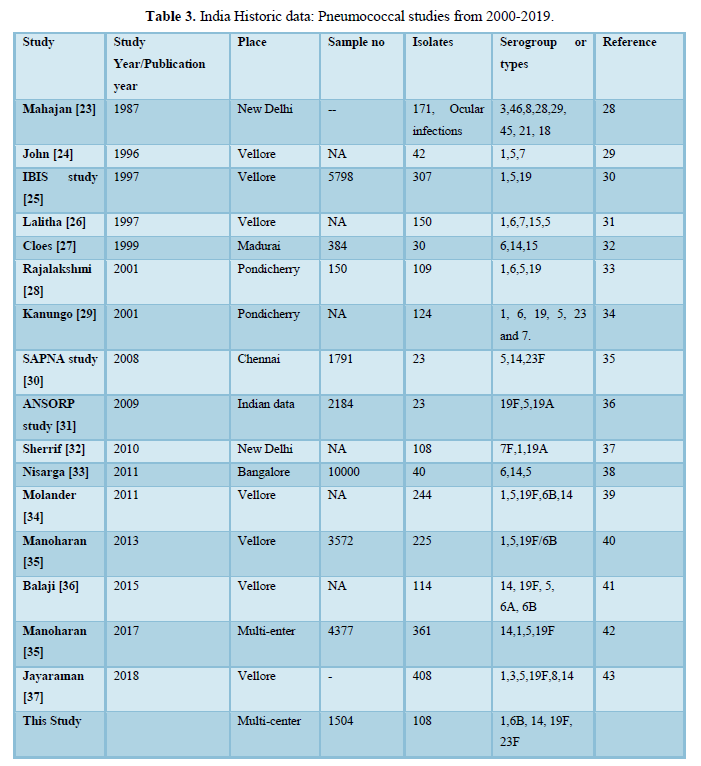
The analysis of Indian publications since 1999 related to invasive pneumococcal disease serotypes revealed serotype 1 as the most common followed by 6A, 19F, 14, 6B and 5. Studies from South India, have reported 1, 6A, 14, 5 as the most common serotypes (Table 3). A multicenter hospital surveillance study reported 6, 1, 19 as the most prevalent serotypes among children

With the characterization of the cps locus of 92 serotypes [38], Leung [8], have recognized a region unique in sequence to all serotypes that is flanked by conserved primer binding sites. Recently diverse groups [5,39,40] have published their results using sequetyping assay as described by Leung [8]. The limitations of Sequetyping method were addressed in the present study.
This study evaluated 1504 serum samples from pediatric patients who had been diagnosed with invasive pneumococcal disease (IPD) from January 2013 to December 2015. High positivity rate for qmPCR (~30%) was observed as compared to conventional methods for isolation of pneumococci (7%). The positivity rates were consistent across different centers of India. Further we were able to deduce serotypes of all 456 qmPCR positive serum samples using PCRSeqTyping. Our study demonstrates the dynamics of diverse serotype distribution of pneumococci in clinical specimens.
PCRSeqTyping based serotyping in PCR positive serum samples indicated the existence geographical variations of serotypes in regions of India. Serotype 1 was observed to predominant in northern part of India and the % distribution was reduced down south. Similarly, serotype was predominant in Southern part of India and its % distribution reduced up north. Few serotypes very uniquely represented in some part of the country. Serotype 9V, 7F, 9N, 23F, 3&4 were found uniquely in Delhi, Jodhpur, Bangalore, Chennai and Kanpur region. Overall serotype 1, 6B, 14, 19F and 23F were predominant serotypes reported from serum samples. This study also demonstrates that pneumococcal serotype can be obtained in cases where is culture is negative. Similar studies were conducted where pneumococcal serotypes were deduced with the aid of molecular serotyping methods directly from blood (201), pleural fluids (202), NP specimens (203), CSF (204) and MEF samples (205). These studies adopted either conventional sequential PCR (93) or real time multiplex sequential PCR (206) for deducing the serotypes from culture negative clinical samples. The major limitation of the studies was that the serotypes of the closely related strains were reported as serogroup. PCRSeqTyping protocol, identified the pneumococcal serotypes directly from serum samples and also assigned the closely related serotypes to their respective types without the issues of cross reaction. The development of modified typing method has the potential to overcome some of the difficulties associated with the conventional serologic method and multiplex/sequential PCR based methods.
Advantages and limitations of PCRSeqTyping
The described method has multiple advantages over other previously reported serotyping methods. It involves techniques with a workflow that many microbiology laboratories can easily implement. The high throughput PCRSeqTyping method features good discriminatory power, reproducibility, and portability, making it suitable for epidemiological studies. The assay has the flexibility of incorporating additional primers for the characterization of emerging serotypes. An added advantage of this method is that raw data from experiments can be reanalyzed upon the addition of new entries to the serotyping database. The limitation of the protocol lies in its inability to quantify the serotype abundance and serotype identification of multiple serotypes commonly encountered in NP samples.
CONCLUSION
Invasive pneumococcal infections are underestimated due to lack of sensitivity of conventional blood culture. Serum qmPCR combined with PCRSeqTyping could identify and serotype, pneumococcal infection precisely in culture negative clinical samples, substantially improving the laboratory diagnosis in children with IPD. Use of these methods will help to estimate the true burden of the disease. Extensive use of this methodology will provide true estimate of disease burden and will help in reducing the treatment failures.
-
United Nations Inter-Agency Group for Child Mortality Estimation (UN IGME), ‘Levels & Trends in Child Mortality (2020) Estimates developed by the United Nations Inter-Agency Group for Child Mortality Estimation’, United Nations Children’s Fund, New York.
-
Farooqui H, Jit M, Heymann DL, Zodpey S (2015) Burden of Severe Pneumonia, Pneumococcal Pneumonia and Pneumonia Deaths in Indian States: Modelling Based Estimates. PLoS One 10(6): e0129191.
-
Jaiswal N, Singh M, Thumburu KK, Bharti B, Agarwal A, et al. (2014) Burden of invasive pneumococcal disease in children aged 1 month to 12 years living in South Asia: A systematic review. PLoS One 9(5): e96282.
-
Vernet G, Saha S, Satzke C, Burgess DH, Alderson M, et al. (2011) Laboratory-based diagnosis of pneumococcal pneumonia: State of the art and unmet needs. Clin Microbiol Infect 3: 1-13.
-
Jin P, Wu L, Oftadeh S, Kudinha T, Kong F, et al. (2016) Using a practical molecular capsular serotype prediction strategy to investigate Streptococcus pneumoniae serotype distribution and antimicrobial resistance in Chinese local hospitalized children. BMC Pediatr 16: 53.
-
Nagaraj G, Ganaie F, Govindan V, Ravikumar KL (2017) Development of PCRSeqTyping-a novel molecular assay for typing of Streptococcus pneumoniae. Pneumonia (Nathan) 9: 8.
-
Ganaie FA, Govindan V, Ravi Kumar KL (2015) Standardization and evaluation of a quantitative multiplex real-time PCR assay for the rapid identification of Streptococcus pneumoniae. Pneumonia (Nathan) 6: 57-66.
-
Leung MH, Bryson K, Freystatter K, Pichon B, Edwards G, Charalambous BM, et al. (2012) Sequetyping: Serotyping Streptococcus pneumoniae by a single PCR sequencing strategy. J Clin Microbiol 50: 2419-2427.
-
Lawrence ER, Arias CA, Duke B, Beste D, Broughton K, (2000) Evaluation of serotype prediction by cpsA-cpsB gene polymorphism in Streptococcus pneumoniae. J Clin Microbiol 38(4): 1319-1323.
-
Reller LB, Weinstein MP, Werno AM, Murdoch DR (2008) Laboratory Diagnosis of Invasive Pneumococcal Disease. Clin Infect Dis 6(15): 926-932.
-
Yildirim I, Shea KM, Pelton SI (2015) Pneumococcal Disease in the Era of Pneumococcal Conjugate Vaccine. Infect Dis Clin North Am 29(4): 679-697.
-
Lawrence ER, Griffiths DB, Martin SA, George RC, Hall LM (2003) Evaluation of semiautomated multiplex PCR assay for determination of Streptococcus pneumoniae serotypes and serogroups. J Clin Microbiol 41: 601-607.
-
Brito DA, Ramirez M, de Lencastre H (2003) Serotyping Streptococcus pneumoniae by multiplex PCR. J Clin Microbiol 41: 2378-2384.
-
Kong F, Gilbert GL (2003) Using cpsA-cpsB sequence polymorphisms and serotype-/group specific PCR to predict 51 Streptococcus pneumoniae capsular serotypes. J Med Microbiol 52: 1047-1058.
-
Kong F, Wang W, Tao J, Wang L, Wang Q, et al. (2005) A molecular-capsular type prediction system for 90 Streptococcus pneumoniae serotypes using partial cpsA-cpsB sequencing and wzy- or wzx-specific PCR. J Med Microbiol 54: 351-356.
-
Pai R, Gertz RE, Beall B (2006) Sequential multiplex PCR approach for determining capsular serotypes of Streptococcus pneumoniae isolates. J Clin Microbiol 44: 124-131.
-
Song JY, Nahm MH, Moseley MA (2013) Clinical implications of pneumococcal serotypes: invasive disease potential, clinical presentations, and antibiotic resistance. J Korean Med Sci 28(1): 4-15.
-
Hanquet G, Kissling E, Fenoll A, George R, Lepoutre A, et al. (2010) Pneumococcal serotypes in children in 4 European countries. Emerg Infect Dis 16(9): 1428-1439.
-
Wahl B, O'Brien KL, Greenbaum A, Majumder A, Liu L, et al. (2018) Burden of Streptococcus pneumoniae and Hemophilus influenzae type b disease in children in the era of conjugate vaccines: global, regional, and national estimates for 2000-2015. Lancet Glob Health 6(7): e744-e757.
-
Kanungo R (2015) Knowledge of serotype prevalence & burden of invasive pneumococcal disease: A prerequisite to vaccine introduction in the country. Indian J Med Res 142(3): 241-244.
-
Manoharan A, Jayaraman R (2018) Pneumococcal vaccines. Indian J Med Microbiol 36(4): 465-474.
-
Jaiswal N, Singh M, Das RR, Jindal I, Agarwal A, et al. (2014) Distribution of serotypes, vaccine coverage, and antimicrobial susceptibility pattern of Streptococcus pneumoniae in children living in SAARC countries: A systematic review. PLoS One 9(9): e108617.
-
Mahajan VM, Bareja U, Prakash K, Ghose S (1987) Pneumococci in ocular disease of children and their treatment. Ann Trop Pediatr 7(4): 270-273.
-
John TJ, Pai R, Lalitha MK, Jesudason MV, Brahmadathan KN, et al. (1996) Prevalence of pneumococcal serotypes in invasive diseases in southern India. Indian J Med Res 104: 205-207.
-
Invasive Bacterial Infection Surveillance (IBIS) Group International Clinical Epidemiology Network (INCLEN) (1999) Prospective multicenter hospital surveillance of Streptococcus pneumoniae disease in India. Lancet 353(9160): 1216-1221.
-
Lalitha MK, Manayani DJ, Priya L, Jesudason MV, Thomas K, et al. (1997) E test as an alternative to conventional MIC determination for surveillance of drug resistant Streptococcus pneumoniae. Indian J Med Res 106: 500-503.
-
Coles CL, Kanungo R, Rahmathullah L, Thulasiraj RD, Katz J, et al. (2001) Pneumococcal nasopharyngeal colonization in young south Indian infants. Pediatr Infect Dis J 20(3): 289-295.
-
Rajalakshmi B (2001) Bacterial pneumonia with special reference to pneumococcal pneumonia: A study of immunological method of diagnosis. Ph.D. Thesis.
-
Kanungo R, Rajalakshmi B (2001) Serotype distribution and antimicrobial resistance in S. pneumoniae causing invasive and other infections in south India. Indian J Med Res 114: 127-132.
-
Thomas K, Steinhoff MC, Lalitha MK, Aparna SS, Sharma P, et al. (2006) Invasive pneumococcal disease in South Asia SAPNA network. In proceedings of the fifth International Symposiuym on Pneumococci and Pneumococcal Diseases, Alice springs, Australia.
-
Song JH, Jung SI, Ko KS, Kim NY, Son JS, et al. (2004) High prevalence of antimicrobial resistance among clinical S. pneumoniae isolates in Asia (as ANSORP study). Antimicrob Agents Chemother 48: 2101-2107.
-
Shariff M, Choudhary J, Zahoor S, Deb M (2013) Characterization of S. pneumonia isolates from India with special reference to their sequence types. J Infect Dev Ctries 7: 101-109.
-
Nisarga R, Premalatha R, Shivananda, Ravikumar KL, Shivappa U, et al. (2015) Hospital based surveillance of Invasive Pneumococcal Disease and Pneumoniae in South Bangalore, India. Indian Pediatr 52: 205-211.
-
Molander V, Elisson C, Balaji V, Backhaus E, John J, et al. (2013) Invasive pneumococcal infections in Vellore, India: Clinical characteristics and distribution of serotypes. BMC Infect Dis 13: 532.
-
Manoharan A, Manchanda V, Balasubramanian S, Lalwani S, Modak M, et al. (2017) Invasive pneumococcal disease in children aged younger than 5 years in India: A surveillance study. Lancet Infect Dis 17: 305-312.
-
Balaji V, Jayaraman R, Verghese VP, Baliga PR, Kurien T (2015) Pneumococcal serotypes associated with invasive disease in under five children in India & implications for vaccine policy. Indian J Med Res 142(3): 286-292.
-
Jayaraman R, Varghese R, Kumar JL, Neeravi A, Shanmugasundaram D, et al. (2018) Invasive pneumococcal disease in Indian adults: 11 years' experience. J Microbiol Immunol Infect 52(5):736-742.
-
Bentley SD, Aanensen DA, Mavroidi A, Saunders D, Rabbinowitsch E, et al. (2006) Genetic analysis of the capsular biosynthetic locus from all 90 pneumococcal serotypes. PLoS Genet 2(3): e31.
-
Dube FS, Mens SP, Robberts L, Wolter N, Nicol P, et al. (2015) Comparison of a Realtime Multiplex PCR and Sequetyping Assay for Pneumococcal Serotyping. PLoS One 10(9): e0137349.
-
Satzke C, Dunne EM, Porter BD, Klugman KP, Mulholland EK (2015) The PneuCarriage Project: A Multi-Centre Comparative Study to Identify the Best Serotyping Methods for Examining Pneumococcal Carriage in Vaccine Evaluation Studies. PLoS Med 12(11): e1001903.
QUICK LINKS
- SUBMIT MANUSCRIPT
- RECOMMEND THE JOURNAL
-
SUBSCRIBE FOR ALERTS
RELATED JOURNALS
- Advance Research on Alzheimers and Parkinsons Disease
- International Journal of Internal Medicine and Geriatrics (ISSN: 2689-7687)
- Journal of Allergy Research (ISSN:2642-326X)
- International Journal of Medical and Clinical Imaging (ISSN:2573-1084)
- Journal of Otolaryngology and Neurotology Research(ISSN:2641-6956)
- Journal of Psychiatry and Psychology Research (ISSN:2640-6136)
- Journal of Blood Transfusions and Diseases (ISSN:2641-4023)





